Bowman Research Highlighted on Cover of Angewandte Chemie
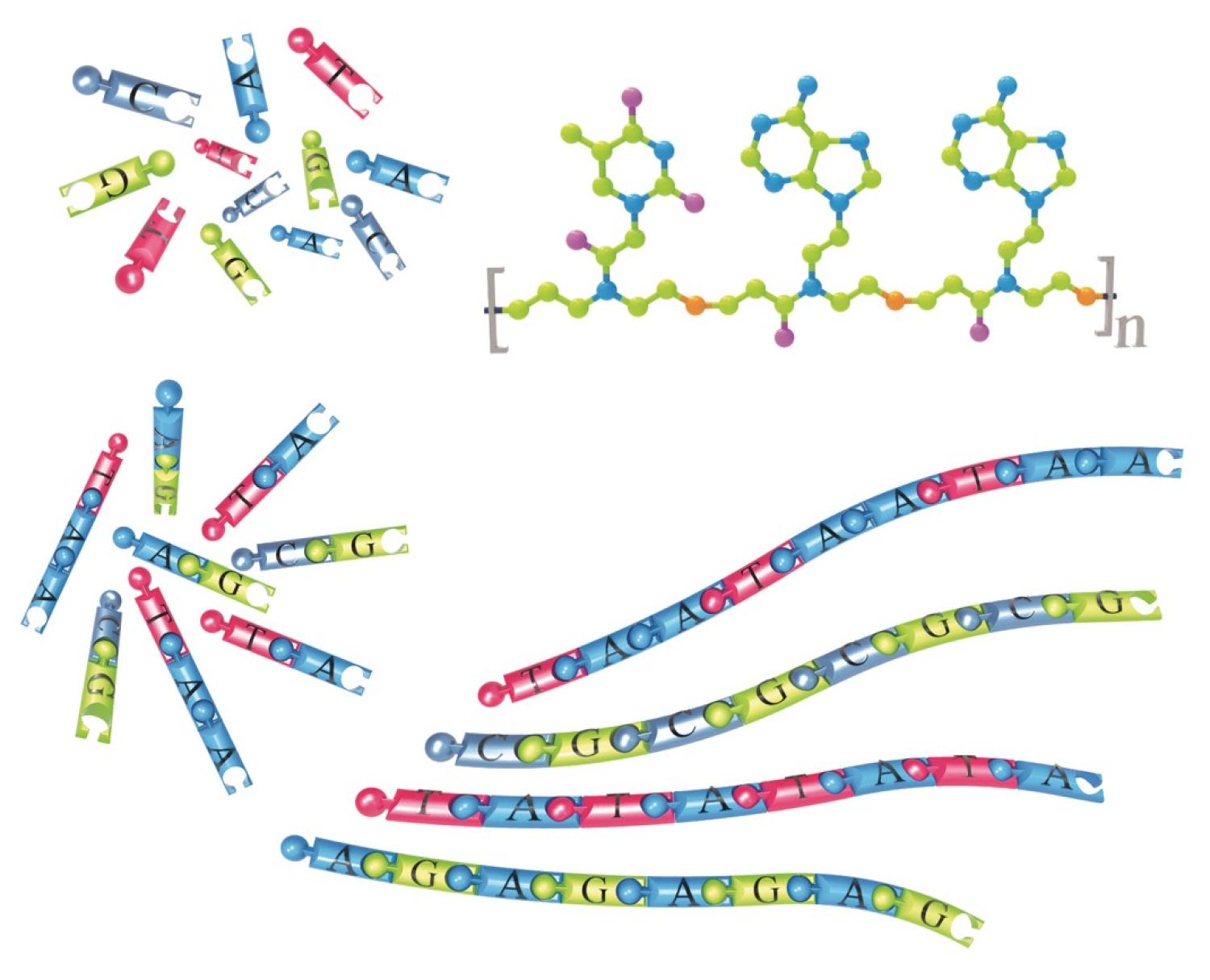
In nature, sequence controls function; differences between you and another person – or organism – primarily arise not from different average DNA compositions, but from different DNA sequences.
Professor Chris Bowmanand his colleagues are seeking to make synthetic polymers with this same capacity, where the sequence itself dictates the properties and behavior. Their work was recently highlighted on the cover of Angewandte Chemie.
“Over the last 50+ years, synthetic polymers, i.e. plastics, have changed the world,” says Bowman, a Distinguished Professor of Chemical and Biological Engineering at the University of Colorado Boulder. “However, the polymers that are used as plastics, as compared to natural polymers such as DNA and proteins, still lack the ability to achieve certain properties because of our inability to control the building block sequence. In contrast, in natural polymers such as DNA, the sequence of the building blocks is precisely defined and controlled. For instance, the specific A, T, G and C nucleobase sequence in DNA polymers is the blueprint for either having a genetic disease or making a functional protein. Moreover, DNA has the ability to recognize complementary sequences of DNA that result in the formation of a duplex.”
Now, Bowman and his colleagues have found a way to create periodically sequence-controlled synthetic polymers that have the power of recognition of complementary sequences like DNA does. They utilize “click” reactions, so called because of their simplicity in clicking molecules together. The process is two-stage. First, a thiol-Michael addition reaction is used to assemble mini-sequences of 2 or 3 nucleobases nearly identical to those in DNA. Subsequently, they use a thiol-ene reaction to link all these mini-sequences together into a longer polymer chain that then has the desired periodic DNA sequence.
“Click reactions are powerful in that they are rapid, high-yielding and robust in nature,” says Bowman. “Moreover, the approach of using one click reaction to first assemble a mini-sequence and then a second to form a polymer with that mini-sequence repeated over and over has much broader applications than just mimicking DNA. For instance, peptide mimics and antimicrobials could be created in this same manner.”
Because their strategy is based on solution phase synthesis, it can be scaled up easily and quickly. The group plans to develop two broadly different types of materials in the long term.
“First, along with numerous collaborators we are targeting nanotechnology applications including manipulation of optical, electronic and mechanical performance of various nanoscale assemblies,” explains Bowman. “Secondly, we are seeking to interact with DNA in biological systems to ultimately develop a new class of biologically active oligonucleotides that could be used, for example, in gene silencing for diseases like Huntington’s or other neurodegenerative diseases.”
Huntington’s disease and other similar neurodegenerative diseases arise because of abnormal and excess repeating mini-sequences in the faulty gene; these mini-sequences are of the same character as Bowman’s synthetic polymer mini-sequences.
“Treatment of some of these types of diseases could be enabled by the delivery of antisense oligonucleotides with similar complementary and repeating sequences,” says Bowman.
In addition to his roles as ChBE Distinguished Professor and the Director of CU-Boulder’s Materials Science and Engineering Program, Bowman is the Associate Director of CU’s Soft Materials Research Center (SMRC). This SMRC recently obtained a $12 million NSF grant to advance soft materials in the areas of click nucleic acids and liquid crystals. Bowman also recently won the 2015 International Association for Dental Research Peyton-Skinner Award for Innovation in Dental Materials.
Learn more:
>> Bowman’s article in Angewandte Chemie
>> $12M Soft Materials Center: Creating DNA Analogs

