Research
Chaperone-mediated Proteasome Assembly
1. How do chaperones mediate "nucleotide-dependent switches" to complete proteasome holoenzymes properly?
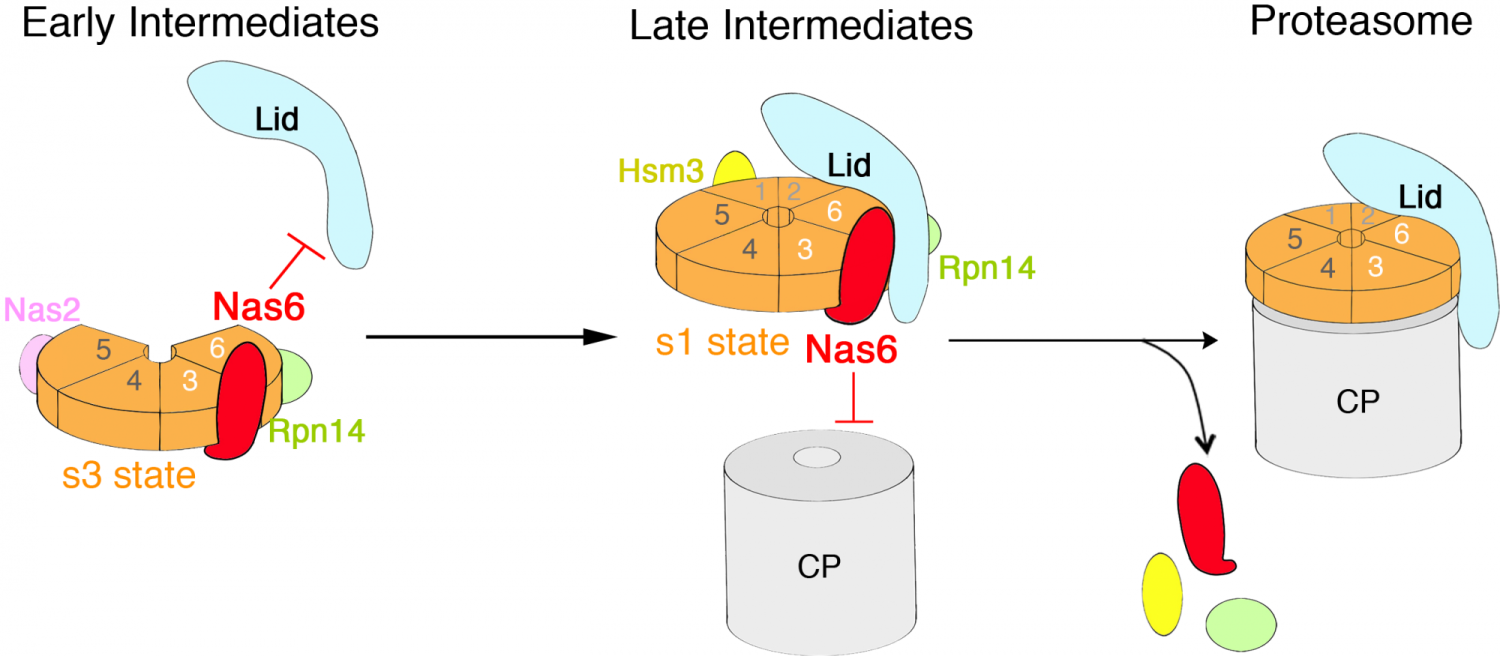
Chaperone-mediated proteasome assembly is an incredibly sophisticated process, which initiates with 2 chaperones (Nas6, Rpn14; red, green) and their cognate ATPase subunits, Rpt3 and Rpt6 (orange). We identified nucleotide-dependent activities of chaperones, using Nas6 (Gankyrin, onco-protein in humans). Nas6 alternates its steric hindrance against lid or CP, depending nucleotide-state of the bound ATPase (orange), until a heterohexameric ATPase ring assembly (orange) is complete. Currently, we are investigating whether and how Nas6 cooperatively acts with its neighboring chaperone, Rpn14 (green), in order to properly "switch" to late-stage intermediates, and to the final step of completing proteasome holoenzyme.
Ubiquitin-dependent Switch
2. How does a "ubiquitin-dependent switch" prevent formation of faulty proteasome holoenzymes?
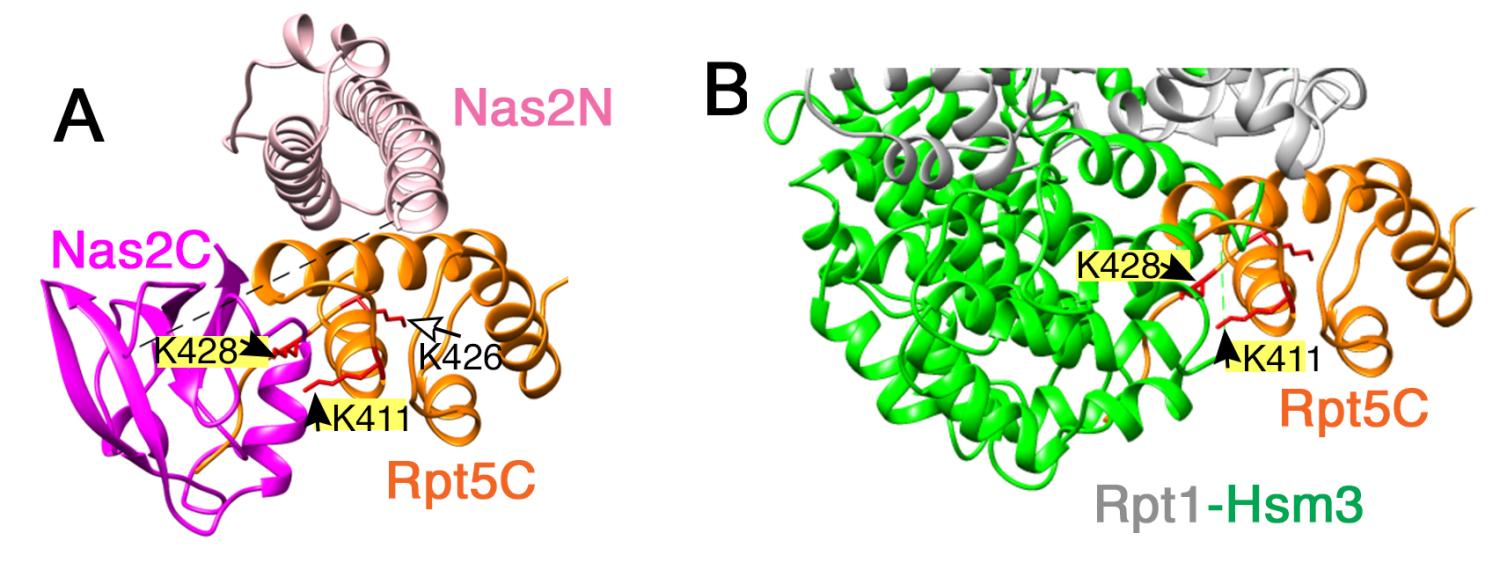
First, we are investigating how Nas2 and Hsm3 act on this penultimate step of a heterohexameric ATPase ring assembly, using a heterologous E.coli expression system in vitro, since we can readily manipulate start and stop point of assembly. Second, we are investigating an additional mechanism of Not4 on chaperone-mediated proteasome assembly, since Not4 also acts as a part of a highly conserved transcriptional and translational regulator, Ccr4-Not complex. Third, we are investigating the fate of a heterohexameric ATPase ring, which is ubiquitinated by Not4, and how this ubiquitination can be reversibly removed to allow the corrected assembly intermediates to proceed to complete proteasome holoenzyme.
 Proteasome Storage Granule
Proteasome Storage Granule

3. How do cells survive nutriotional stress by "remodeling" proteasomes for storage against autophagy?
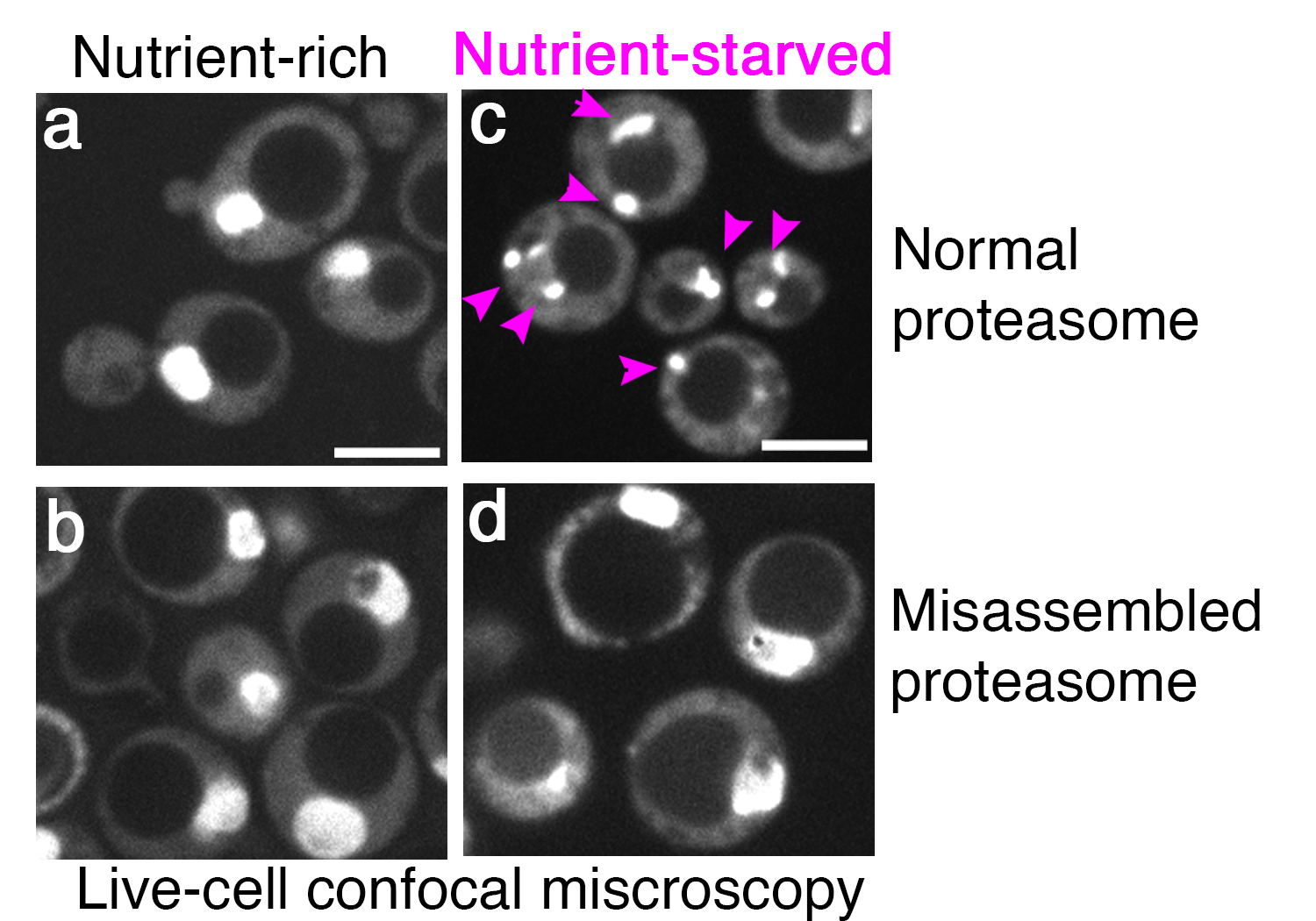
When nutrients are abundant, proteasomes are enriched in the nucleus where they degrade key regulators of fundamental processes, such as transcription factors and cell cycle regulators. By contrast, when nutrients are scarce, proteasomes dramatically remodel, exit the nucleus, and store in cytoplasmic "proteasome storage granule" for protection against autophagy. Only the properly assembled proteasomes can be stored, and misassembled proteasomes cannot. We aim to identify how de novo proteasome assembly via chaperones influences the fate of proteasomes during nutriontional stress, and how cancer cells overexpressing a known onco-protein, Gankyrin, influence proteasome storage and protection against autophagy for their survival, as they encounter nutriotional fluctuations.

