Research
Uncovering the role of transposons in immune gene regulation
Our genomes are littered with parasitic genetic sequences that serve no biological function other than to selfishly replicate themselves within the genome. These are largely considered genetic glitches, yet there is emerging evidence that transposons are an important source of "recycled" genetic parts that can sometimes help their hosts rather than hurt them.
Our research seeks to understand how these "bugs" have influenced our genetic code. We employ both computational and experimental approaches to decipher how transposons impact genome architecture and organismal biology. Our research lies at the crossroads of diverse fields including genomics, epigenetics, evolution, immunity, and disease.
A few of our major research areas are detailed below.
Widespread domestication of transposons for immune gene regulation
Our lab has demonstrated that transposons have been repeatedly co-opted to regulate immune genes. Although different lineages harbor distinct repertoires of transposable elements, we found widespread co-option of transposons as regulatory elements affecting immune responses in multiple species, establishing that transposons facilitate adaptive rewiring of immune regulatory networks.
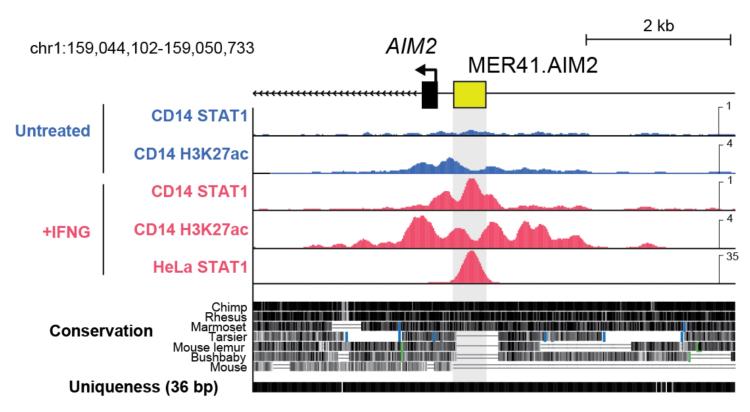
An endogenous retrovirus regulates inducible expression of the antiviral gene AIM2
- Horton I#, Kelly CJ#, et al, Mouse B2 SINE elements function as IFN-inducible enhancers. eLIFE. 2023
- Kelly CJ, et al. Ruminant-specific retrotransposons shape regulatory evolution of bovine immunity. Genome Res. 2022
- Buttler et al, An intronic LINE-1 regulates IFNAR1 expression in human immune cells. Mobile DNA. 2023.
- Chuong EB, et al. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016
Transposon-mediated evolution of new protein isoforms
Most recently, we have begun to explore how transposons influence splicing, leveraging advances in sequence assembly and long-read RNA-seq technology. We have uncovered dozens of cases in multiple species where immune genes express transposon-derived isoforms, and have characterized a subset of these examples. In our latest study we characterized an isoform of the IFN receptor IFNAR2 as a novel, broadly expressed regulator of human IFN signaling, supporting a significant role for these isoforms in shaping species-specific immunity.
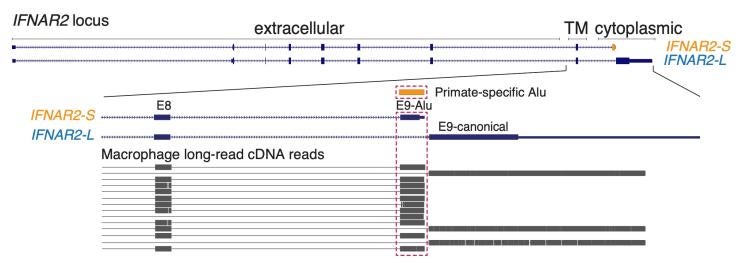
Long-read transcriptomic data showing prominent expression of an IFNAR2 isoform derived from transposon exonization.
- Pasquesi et al, Transcriptional dynamics of transposable elements in the type I IFN response in Myotis lucifugus cells. Mobile DNA. 2022
Pasquesi et al, Regulation of human interferon signaling by transposon exonization. Cell. 2024
Consequences of transposon dysregulation in cancer
It is increasingly appreciated that transposon reactivation has many pathological consequences in cancer, including gene dysregulation. We discovered that some transposons act as tumor-specific regulatory elements and mediate transcriptional rewiring in colorectal cancer cells.
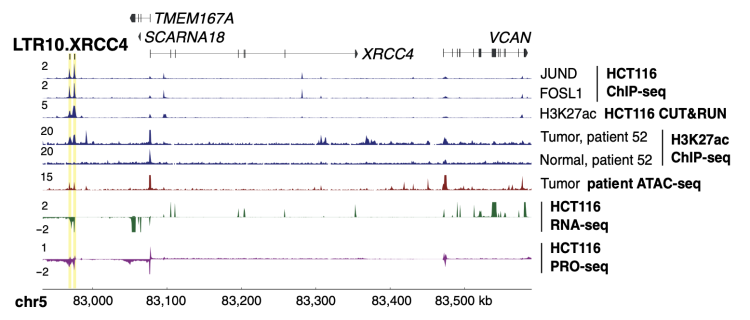
- Ivancevic et al, Endogenous retroviruses mediate transcriptional rewiring in response to oncogenic signaling in colorectal cancer, Science Advances. 2024
- Dziulko et al, An endogenous retrovirus regulates tumor-specific expression of the immune transcriptional regulator SP140. Human Molecular Genetics. 2024

