Antibiotic Effectiveness in Space (AES-1)
Funding Agencies
Dates
- Launched to space on Orbital CRS-1, January 2014
- Operated in Space by: NASA astronauts Rick Mastracchio and Mike Hopkins on board the International Space Station (Expedition 38)
- Returned to Earth: SpaceX CRS-3 (May, 2014) and SpaceX CRS-4 (October, 2014)
Previous experiments conducted in space have shown that under certain conditions, bacteria can grow to larger numbers, need more drugs to be killed, and can have an increased ability to cause disease in space with respect to how they behave on Earth. Why is this? Can we use this knowledge in our fight against drug-resistant bacteria on Earth?
What was this project’s objective?
The main objective of the AES-1 experiment was to determine changes that can be measured under the microscope (phenotypic) like cell size, cell envelope thickness, and final cell count, and those that happen at a molecular genetic level (gene expression) that allow Escherichia coli to survive in space in concentrations of antibiotics that on Earth would kill them.
How was this done?
To achieve this, sets of E. coli were prepared inside BioServe Space Technologies’ Fluid Processing Apparatus (FPA) and Group Activation Packs (GAP). NASA astronauts Rick Mastracchio and Mike Hopkins operated the GAPs in space, while the matching controls on Earth were operated at BioServe Space Technologies in Boulder, Colorado. Samples were challenged with an antibiotic called Gentamicin Sulfate. A total of 128 samples were launched to space and another 128 equivalent samples were cultured on Earth.

NASA Astronaut Rick Mastracchio shown holding a GAP in his right hand and the crank used to conduct an activation in the other as he performed the experimental operations on board the International Space Station. The open CGBA incubator loaded with other 15 GAPs is in the bottom right. Image: NASA. From: Zea L., Larsen M, Estante F, Qvortrup K, Moeller R, Dias de Oliveira S, Stodieck L and Klaus D, (2017) Phenotypic Changes Exhibited by E. coli Cultured in Space. Front. Microbiol. 8:1598. doi: 10.3389/fmicb.2017.01598
Results
Cell size: it was found that E. coli cells cultured in space were 59% the length, and 83% the diameter of their controls on Earth. This means that, in space, bacteria grew to be only 37% the volume of their Earth controls.
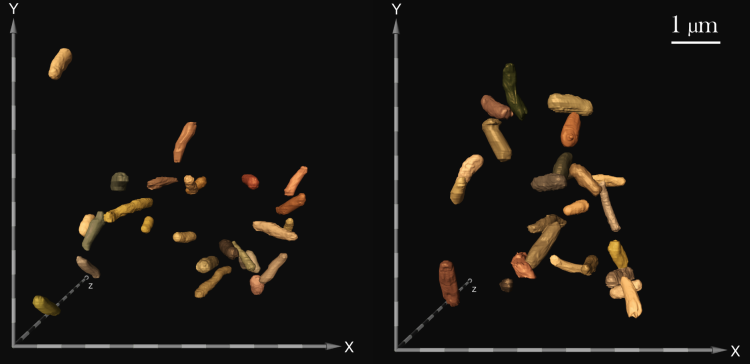
In space, E. coli cells were 59% the length, 83% the diameter, and thus 37% the volume of Earth controls. Focused Ion Beam/Scanning Electron Microscopy (FIB/SEM) image of samples challenged with 25 µg/mL gentamicin: Spaceflight (left) and Earth control (right). FIB/SEM analysis corroborated the findings on reduced cell size in space. Scale bar on axes = 500 nm. Image produced by the Department of Biomedical Sciences, University of Copenhagen. From: Zea L., Larsen M, Estante F, Qvortrup K, Moeller R, Dias de Oliveira S, Stodieck L and Klaus D, (2017) Phenotypic Changes Exhibited by E. coli Cultured in Space. Front. Microbiol. 8:1598. doi: 10.3389/fmicb.2017.01598
Cell envelope thickness and ultrastucture: Cell envelopes in space were between 25% and 43% thicker than on Earth. Some of the bacteria that grew in space in concentrations of antibiotic that on Earth would kill them, showed irregular shapes and the formation of ‘outer membrane vesicles’, capsules that help bacteria communicate with one another.
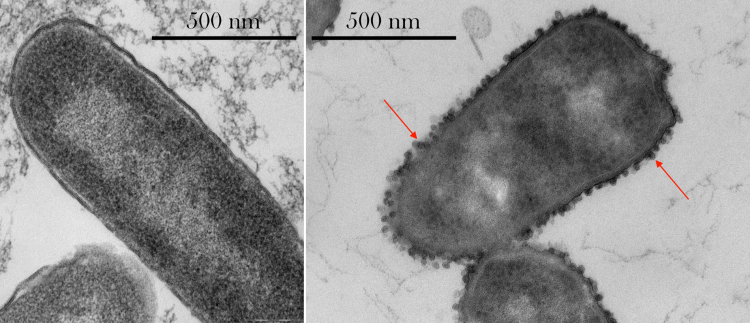
Thin-section Transmission Electron Microscopy (TEM) images of E. coli. Left: sample cultured on Earth and challenged with 50 µg/mL gentamicin. Right: sample cultured in space and challenged with 175 µg/mL shows the presence of extracellular vesicles (red arrows) and irregular cellular shapes. Images taken with a Philips CM 100 TEM at an accelerating voltage of 80 kV. Image produced by the Department of Biomedical Sciences, University of Copenhagen. From Zea L., Larsen M, Estante F, Qvortrup K, Moeller R, Dias de Oliveira S, Stodieck L and Klaus D, (2017) Phenotypic Changes Exhibited by E. coli Cultured in Space. Front. Microbiol. 8:1598. doi: 10.3389/fmicb.2017.01598
Final cell count: At the end of the experiment, in average, for every bacterium there was on Earth, there were 13 in the samples cultured in space. More details on this, here: Zea L, Larsen M, Estante F, Qvortrup K, Moeller R, Dias de Oliveira S, Stodieck L and Klaus D (2017) Phenotypic Changes Exhibited by E. coli Cultured in Space. Front. Microbiol. 8:1598. doi: 10.3389/fmicb.2017.01598
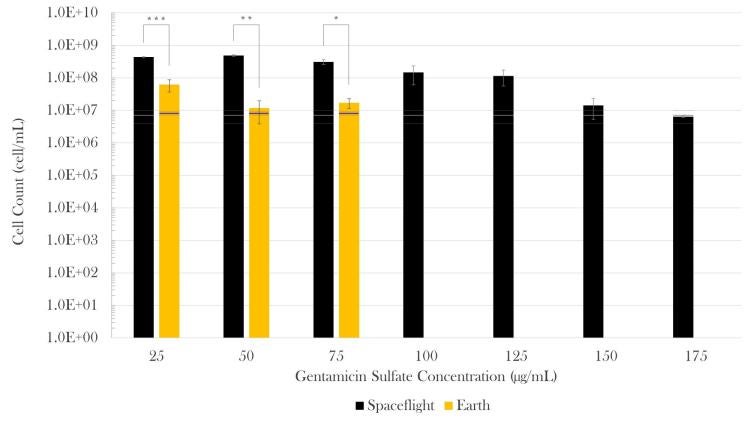
E. coli growth challenged with gentamicin sulfate. Black bars represent spaceflight; gold bars are Earth samples. The gray and blue lines indicate the cell concentration at the time of antibiotic introduction in space and Earth, respectively, showing the average (center line) and standard errors (upper and lower lines). Spaceflight cultures showed higher cell counts than their Earth matched controls (13-fold increase in average). Although there appears to be a decrease in magnitude at 150 and 175 µg/mL with respect to 125 µg/mL, this is misleading as accurate values were hard to acquire either by cell count or optical density due to cell aggregation in these samples. It is estimated that values at 150 and 175 μg/mL were roughly equivalent to that of 125 µg/mL. Bars indicate standard error; n = 4 for all except for spaceflight at 25 and 50 µg/mL (n = 3, each), and 175 µg/mL (n = 2), as only the samples for which it was certain that the antibiotic was fully introduced were considered. A star means p<0.05, two indicate p<0.01 and three p<0.001. From Zea L., Larsen M, Estante F, Qvortrup K, Moeller R, Dias de Oliveira S, Stodieck L and Klaus D, (2017) Phenotypic Changes Exhibited by E. coli Cultured in Space. Front. Microbiol. 8:1598. doi: 10.3389/fmicb.2017.01598
Gene expression: genes used by E. coli to defend itself against acidic conditions were ‘turned on’ in space, as well as those that these bacteria use when they are in starving conditions.
What do these results mean?
Studying the gene expression (transcriptomics) allow scientist to understand what may cause the results being observed. In the case of AES-1, the gene expression data indicated that in space cells were starving and in high-acidic conditions. This was surprising, as the cultures on orbit had the same amount of nutrients, and same pH at the end of the experiment as the cultures grown on Earth. This suggested that there was a zone around a cell where nutrients were lacking (as the cell was consuming those food molecules available) and acidic compounds were building up (natural by-products of the bacterial metabolism). This had been hypothesized since the 1980’s (sometimes called an ‘altered extracellular environment in space’) by multiple scientists, but it had not been either confirmed or denied by empirical or computational approaches, and data supporting it was finally obtained by this gene expression study. More details on this, here: Zea, L., Prasad, N., Levy, S., Stodieck, L., Jones, A., Shrestha, S., Klaus, D., (2016). A Molecular Genetic Basis to Bacterial Behavior in Space, PLoS ONE 11(11): e0164359
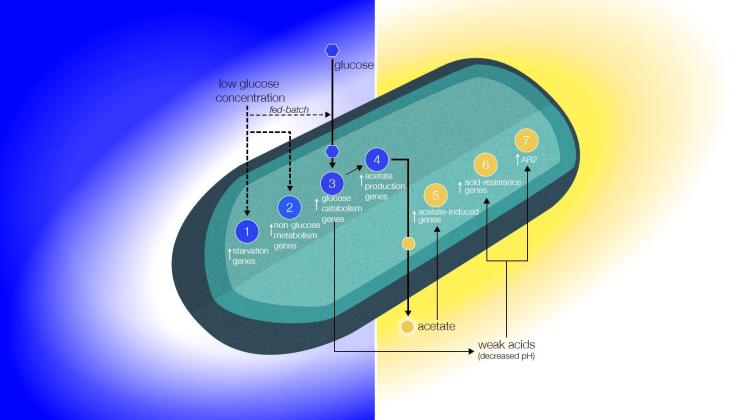
Altered Extracellular Model. Biomolecular model based on the gene expression data analyses support the reduction of glucose molecules (blue gradient) and acid buildup (gold gradient) proposed to occur in the boundary layer around the cell. This altered extracellular environment has been hypothesized to result as an effect of reduced gravity-driven forces acting on the cell-fluid system and has been put forth as the biophysical mechanism governing bacterial behavior in space. Blue circles indicate overexpression of genes associated with metabolism, while gold circles represent the overexpression of acidic condition genes. From: Zea L, Prasad N, Levy SE, Stodieck L, Jones A, Shrestha S, et al. (2016) A Molecular Genetic Basis Explaining Altered Bacterial Behavior in Space. PLoS ONE 11(11): e0164359. doi:10.1371/journal.pone.0164359
Another interesting finding was that genes that allow bacteria to become resistant to antibiotics were turned on in space but not directly. The increase in the acidity around the cell caused several acidity-defense genes to be turned on and these, in turn, activated mechanisms in the bacteria that allow it to survive higher concentrations of drugs. More details on this, here: Aunins, T. R., Erickson, K. E., Prasad, N., Levy, S. E., Jones, A., Shrestha, S., Mastracchio, R., Stodieck., L., Klaus, D., Zea, L., Chatterjee, A. (2018). Spaceflight Modifies Escherichia coli Gene Expression in Response to Antibiotic Exposure and Reveals Role of Oxidative Stress Response. Frontiers in Microbiology, 9, 310
Project Bibliography
Ph.D. Thesis
- Zea, L., Phenotypic and Gene Expression Responses of E. coli to Antibiotics during Spaceflight, Ph.D. Thesis, University of Colorado, Boulder, CO, 2015
Journal Articles
- Aunins, T. R., Erickson, K. E., Prasad, N., Levy, S. E., Jones, A., Shrestha, S., Mastracchio, R., Stodieck., L., Klaus, D., Zea, L., Chatterjee, A. (2018). Spaceflight Modifies Escherichia coli Gene Expression in Response to Antibiotic Exposure and Reveals Role of Oxidative Stress Response. Frontiers in Microbiology, 9, 310
Zea L., Larsen M, Estante F, Qvortrup K, Moeller R, Dias de Oliveira S, Stodieck L and Klaus D, (2017) Phenotypic Changes Exhibited by E. coli Cultured in Space. Front. Microbiol. 8:1598. doi: 10.3389/fmicb.2017.01598
Zea, L., Prasad, N., Levy, S., Stodieck, L., Jones, A., Shrestha, S., Klaus, D., (2016). A Molecular Genetic Basis to Bacterial Behavior in Space, PLoS ONE 11(11): e0164359
Conference Papers, Abstracts, and Posters
Zea, L., Stodieck, L. and Klaus, D., Characterizing Phenotypic and Gene Expression Changes in E. coli Challenged with Antibiotics during Spaceflight, 4th International Space Station Research and Development Conference, Boston, MA, July 7-9, 2015
Zea, L., Stodieck, L. and Klaus, D., Preliminary Results of the Antibiotic Effectiveness in Space-1 (AES-1) Experiment Conducted Onboard ISS, ASGSR Conference, Pasadena, CA, October 22-26, 2014
Zea, L., Stodieck, L., Klaus, D.M., Bacterial Growth and Susceptibility to Antibiotics in Simulated Reduced Levels of Gravity, ASGSR Conference, Orlando, FL November 3-8, 2013
Links
NASA story: https://www.nasa.gov/mission_pages/station/research/news/aes_1/
NASA webpage: https://www.nasa.gov/mission_pages/station/research/experiments/1165.html
CASIS Upward magazine: https://upward.iss-casis.org/microbes-in-microgravity/
NASA video: https://www.youtube.com/watch?v=GeBqMANGtmI
NASA Pre-Launch Science Briefing video: https://www.youtube.com/watch?v=js-AfIbeuW4

