Research
Understanding evolution of antimicrobial resistance using Systems & Computational Biology approaches
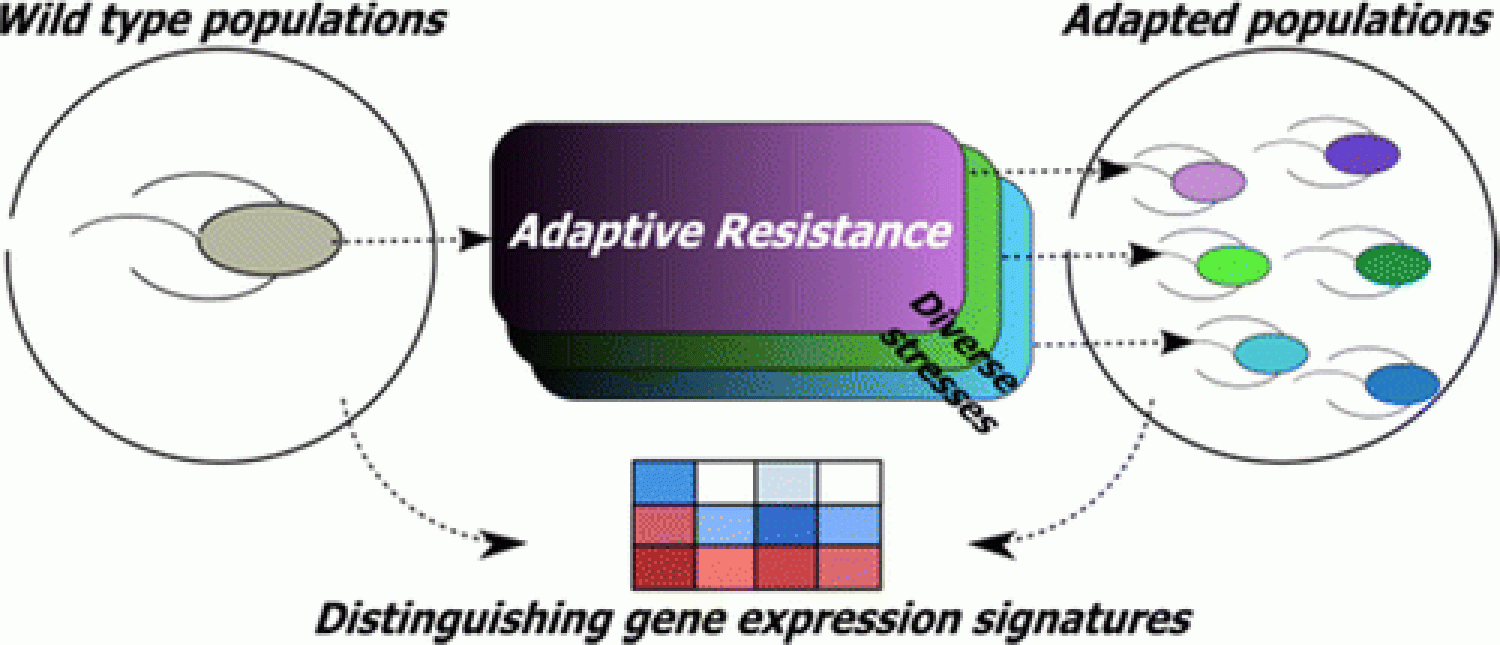
Antibiotics have been an important weapon for treating bacterial infections, however, extensive use and misuse of antibiotics has resulted in selection and worldwide spread of antibiotic resistant bacteria, resulting in failure of standard antibiotics-based therapy, causing prolonged illness or even death. In the near future, antimicrobial resistance will be the primary driver of effective antimicrobial drug development activities.
The root cause of the antibiotic resistance crisis is the ability of bacteria to evolve resistance to a multitude of antibiotics and other environmental toxins. The regulation of adaptation is difficult to pinpoint due to extensive phenotypic heterogeneity arising during evolution. We aim at investigating the mechanisms underlying general bacterial adaptation by evolving wild-type and clinical bacterial populations to dissimilar chemical toxins. We investigate extensive inter- and intrapopulation phenotypic heterogeneity, genomic and transcriptomic heterogenity across adapted populations to indentify "Universal signatures" of adaptive resistance. We explore the connection between gene expression variability and adaptation, using single-gene knockout and CRISPR (clustered regularly interspaced short palindromic repeats) interference strains and quantify impact on adaptation to antibiotics. We identify patterns in gene expression as an indicator of bacterial adaptive resistance, even in the face of the pervasive phenotypic heterogeneity underlying adaptation. Using mathematical modeling and molecular biology tools we adopt a multi-pronged strategy to identify novel gene targets that can potentially prevent or slow down emergence of drug-resistance. This research will involve a combination of various disciplines including chemical engineering, systems biology, microbiology, bioinformatics, computational biology and molecular biology, and will foster multi-disciplinary collaborative environment.
Publications:
Keesha E. Erickson, Peter B. Otoupal, and Anushree Chatterjee* (2015), "Gene expression variability underlies adaptive resistance in phenotypically heterogenous bacterial populations." ACS Infectious Diseases 1(11), pp 555-567.
Anushree Chatterjee#, Laura C. Cook#, Che-Chi Shu, Yuching Chen, Dawn Manias, Doraiswami Ramkrishna, Gary M. Dunny and Wei-Shou Hu* (2013), “Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer.” Proceedings of National Academy of Sciences, 110(17), 7086-7090.
Anushree Chatterjee, Christopher M. Johnson, Che-Chi Shu, Yiannis N. Kaznessis, Doraiswami Ramkrishna, Gary M. Dunny and Wei-Shou Hu* (2011).“Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation.” Proceedings of the National Academy of Sciences, 108: 9721-9726.
Che-Chi Shu, Anushree Chatterjee, Gary M. Dunny, Wei-Shou Hu and Doraiswami Ramkrishna* (2011), “Bistability versus Bimodal Distributions in Gene Regulatory Processes from Population Balance Model.” PLoS Computational Biology, 7(8):e1002140.
Laura CC Cook, Anushree Chatterjee, Aaron Barnes, Jeremy Yarwood, Wei-Shou Hu and GaryM. Dunny*(2011), “Biofilm growth alters regulation of conjugation by a bacterial pheromone.” Molecular Microbiology, 81(6), 1499-1510.
Che-Chi Shu,Anushree Chatterjee, Wei-Shou Hu and Doraiswami Ramkrishna* (2012), “Modeling of Gene regulatory processes by Population mediated Signaling. New applications of Population balances.” Chemical Engineering Science, 70: 188-199.
Design of “next-generation smart antimicrobials" Using Synthetic biology, Nanotechnology and Computational Biology approaches
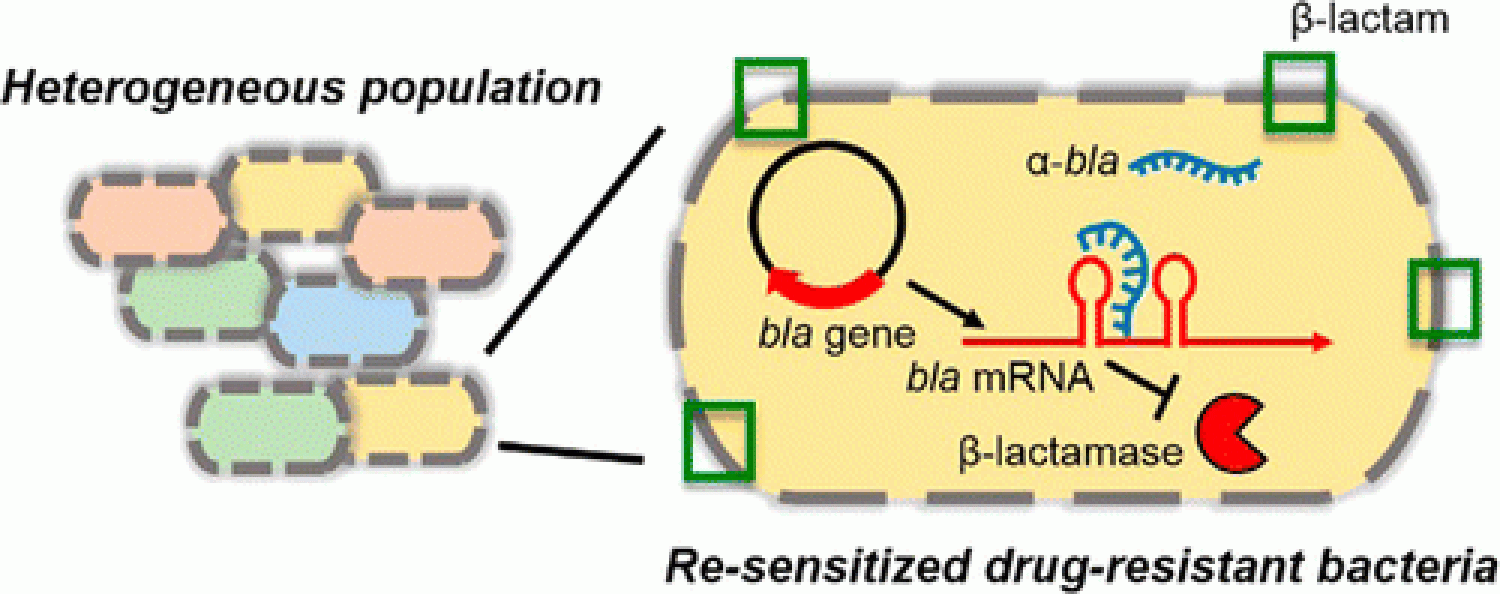
Antibiotics have been an important weapon for treating bacterial infections, however, extensive use and misuse of antibiotics has resulted in selection and worldwide spread of antibiotic resistant bacteria, resulting in failure of standard antibiotics-based therapy, causing prolonged illness or even death. In the near future, antimicrobial resistance will be the primary driver of effective antimicrobial drug development activities.
We aim at developing “next-generation smart antimicrobials” by rationally engineering novel therapeutics that target essential bacterial/viral genes in a potentially resistance-free manner. With the aid of synthetic biology, nanoengineering, mathematical modeling and experimentation we investigate the mutational space and mutational plasticity for resistance evolution. Using biophysical modeling and molecular biology tools we adopt a multi-pronged strategy to design novel antimicrobials that potentially prevent emergence as well as attainment of drug-resistance including nucliec acid therapuetics, CRISPR based antimicrobials, bacteriophages. We also aim at investigating unique therapeutic strateugues including design of metal nanoparticles that can induce cell death specifically in MDR bacteria . Recently, we have shown that photoexcited quantum dots (QDs) can kill a wide range of MDR bacterial clinical isolates, including methicillin-resistant Staphylococcus aureus, carbapenem-resistant Escherichia coli, and extended-spectrum β-lactamase-producing Klebsiella pneumoniae and Salmonella typhimurium. The killing effect is independent of material and controlled by the redox potentials of the photogenerated charge carriers, which selectively alter the cellular redox state. We also show that the QDs can be tailored to kill 92% of bacterial cells in a monoculture, and in a co-culture of E. coli and HEK 293T cells, while leaving the mammalian cells intact, or to increase bacterial proliferation. Our "smart antimicrobial" research will involve a combination of various disciplines including chemical engineering, structural biology, systems biology, microbiology, nanotechnologym computational biology and molecular biology, and will foster multi-disciplinary collaborative environment.
Publications:
Colleen M. Courtney#, Samuel Goodman#, Jessica Mc Daniel, Nancy Madinder, Anushree Chatterjee*, and Prashant Nagpal* (2016), "Photoexcited Quantum Dots for Killing Multi-Drug Resistant Bacteria." Nature Materials. Advance Online Publication. DOI: 10.1038/nmat4542
Colleen M. Courtney, and Anushree Chatterjee* (2015), "Sequence-specific peptide nucleic acid based antisense inhibitors of TEM-1 beta-lactamase and mechanism of adaptive resistance." ACS Infectious Diseases 1 (6), pp 253–263.
Anushree Chatterjee#, Laura C. Cook#, Che-Chi Shu, Yuching Chen, Dawn Manias, Doraiswami Ramkrishna, Gary M. Dunny and Wei-Shou Hu* (2013), “Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer.” Proceedings of National Academy of Sciences, 110(17), 7086-7090.
Anushree Chatterjee, Christopher M. Johnson, Che-Chi Shu, Yiannis N. Kaznessis, Doraiswami Ramkrishna, Gary M. Dunny and Wei-Shou Hu* (2011).“Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation.” Proceedings of the National Academy of Sciences, 108: 9721-9726.
Engineering novel Synthetic genetic devices and Logic gate devices for Higher Order Biological Computation using Experimental and Computational Approaches
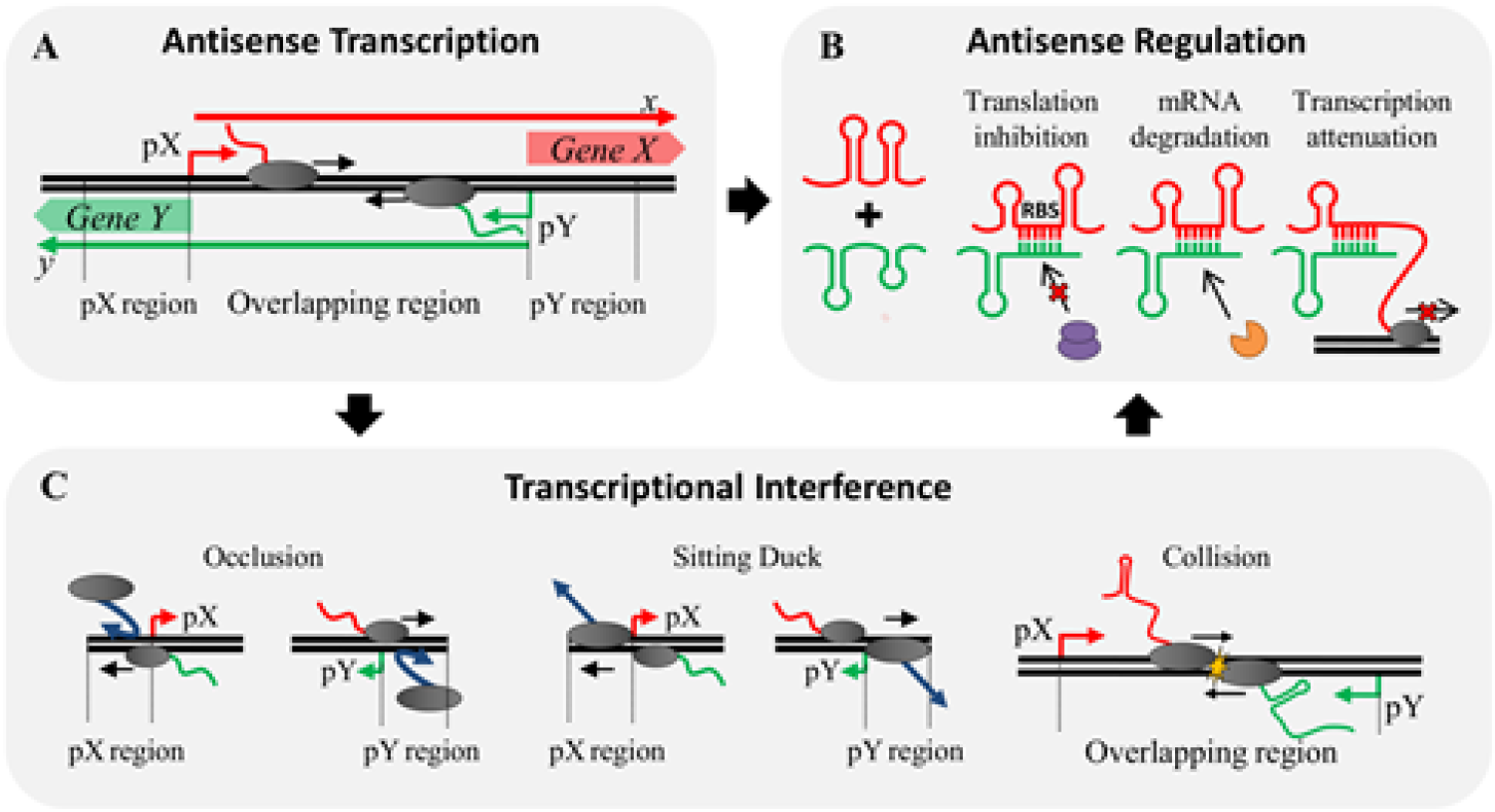
We aim at designing, constructing and engineering modular synthetic genetic devices that process complex environmental inputs into controlled system outputs for a variety of biotechnological, material, medical and bioenergy applications.The research work will include rational design of biological parts such as transcription factors, promoter sequences, receptors, feedback loops, regulatory RNA, and transcriptional traffic to build complex genetic networks to achieve higher-order biological computation. Such synthetic genetic devices would be integrated with complex metabolic networks to optimize cellular machinery for production of desired substances including bio-fuels and pharmaceuticals, and for gene therapy applications.
A major focus of the technology developed would be to apply systems biology approaches to understand and manipulate such metabolic networks. Development of these devices will allow for construction of tight and high-order gene regulatory networks with highly tunable system output such as bistable switches, oscillatory networks etc. The research will involve combining molecular biology, biophysical computation, mathematical modeling, synthetic and systems biology based approaches, and will foster multi-disciplinary collaborative environment.
Publications:
- Antoni E. Bordoy, and Anushree Chatterjee* (2015), "cis-Antisense Transcription Gives Rise to a Tunable Genetic Switch Behavior: A Mathematical Modeling Approach." PLoS ONE 10(7): e0133873. doi:10.1371/ journal.pone.0133873.
- Anushree Chatterjee, Laura C. Cook, Che-Chi Shu, Yuching Chen, Dawn Manias, Doraiswami Ramkrishna, Gary M. Dunny and Wei-Shou Hu* (2013), “Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer.” Proceedings of National Academy of Sciences, 110(17), 7086-7090.
- Anushree Chatterjee, Christopher M. Johnson, Che-Chi Shu, Yiannis N. Kaznessis, Doraiswami Ramkrishna, Gary M. Dunny and Wei-Shou Hu* (2011).“Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation.” Proceedings of the National Academy of Sciences, 108: 9721-9726.
- Colleen M. Courtney and Anushree Chatterjee* (2014), “cis-Antisense RNA and Transcriptional interference: coupled layers of gene regulation.” Journal of Gene Therapy 2(1):9. (Link) (Invited Review)
Sequencing and Diagnostics to detect Superbugs early
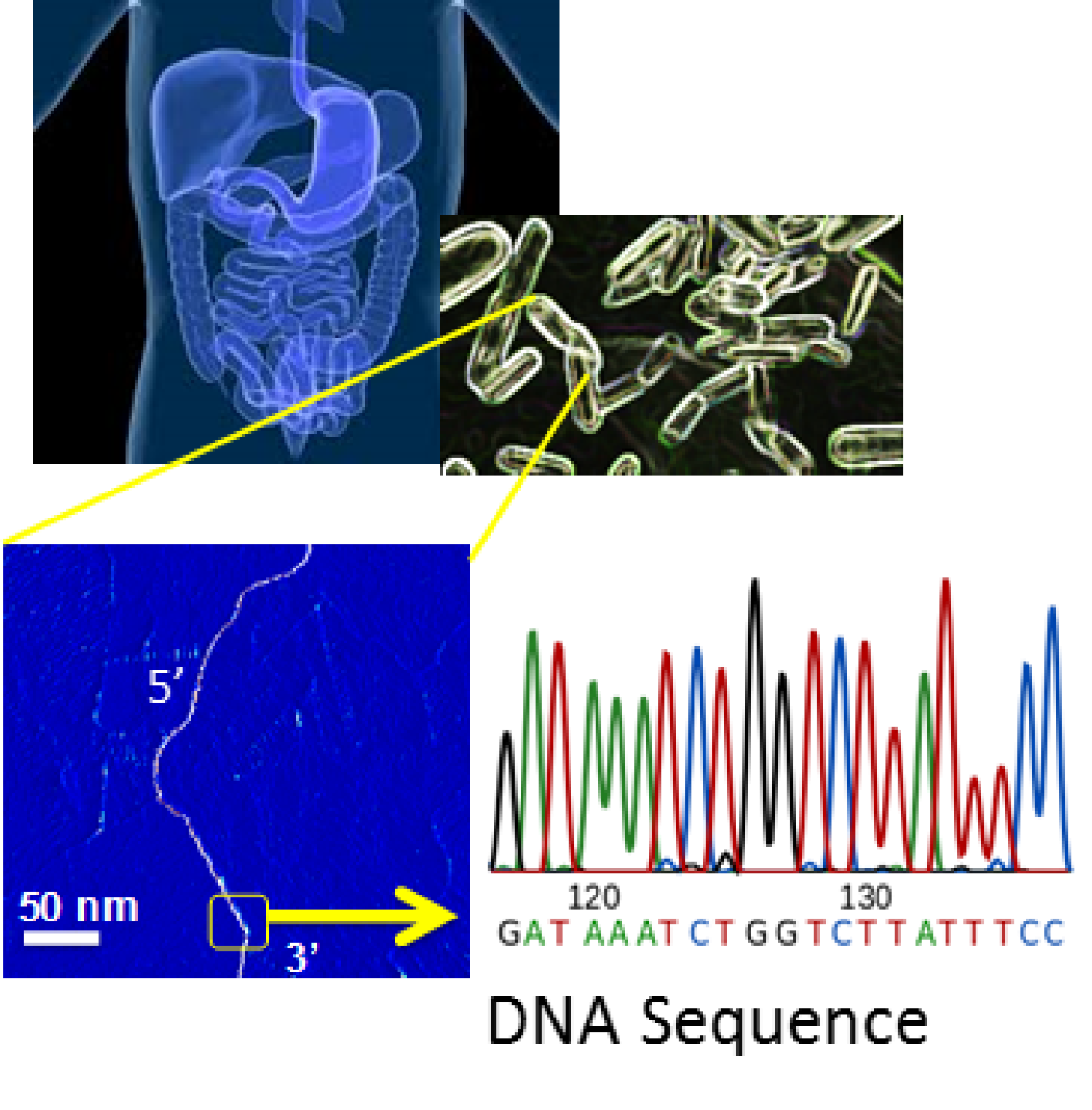
We are developing diagnostic platforms that can be used to detect and sequence trace amounts of drug-resistant variants, with “single-molecule” precision, and with immediate implications in the clinical settings.
Our lab was recently recongnized Quantum Sequencing technology developed by our and Nagpal lab with the New Inventor of the Year award. We are developing a platform technology for fast, reliable, high-throughput and cost effective single molecule sequencing of nulciec acids. This kind of sequencing is an important step in the developement of new diagnostic tools for personalized medicine, as well as in rapid identification of DNA sequences that allow bacteria to develop drug resistance. This approach can potentially transform how we detect drug resistant pathogens in the clinical setting, will allow faster diagnostics and early detection of drug resistant strains that can prevent future spread of resistance, and will also reduce the cost of diagnostics.
The research will involve combining microbiology, molecular biology, biophysical computation, density functional theory, physics, bioinformatics based approaches, and will foster multi-disciplinary collaborative environment.
Publications:
- Josep Casamada Ribot, Anushree Chatterjee*, and Prashant Nagpal* (2015), "Measurements of Single nucleotide Electronic states as Nanoelectronic fingerprints for Identification of DNA Nucleobases, their Protonated and Unprotonated states, Isomers and Tautomers." The Journal of Physical Chemistry Letters B 119 (15), pp 4968–4974.
- Samuel Goodman, Vivek Singh, Josep C. Ribot, Anushree Chatterjee, and Prashant Nagpal* (2014). "Multiple Energy Exciton shelves in Quantum Dot-DNA Nano-bioelectronics." The Journal of Physical Chemistry Letters (5) 5, pp 3909–3913.
Understanding Microbial gene-networks and metabolic networks using Experimental and Computational Approaches
We aim at de-constructing naturally-occurring biologically relevant gene-networks that are essential for bacterial survival, and rationally re-design them bottom-up using library of biological parts to identify key regulatory components that can be used to design new antimicrobials. We are also developing computational and experimental tools to study and optimize metabolic networks for desgning microbial strains for biotechnological applications and drug discovery.
The research will involve combining microbiology, genome engineering metabolic engineering, flux balance analysis, mathematical modeling approaches, and will foster multi-disciplinary collaborative environment.
Publications:
- Antoni E. Bordoy, and Anushree Chatterjee* (2015), "cis-Antisense Transcription Gives Rise to a Tunable Genetic Switch Behavior: A Mathematical Modeling Approach." PLoS ONE 10(7): e0133873. doi:10.1371/ journal.pone.0133873.
- Keesha E Erickson, Ryan T Gill, and Anushree Chatterjee* (2014), "CONSTRICTOR: Constraint modification provides insight into design of biochemical networks." PLoS ONE 9(11):e113820. doi:10.1371/journal.pone.0113820.
- Anushree Chatterjee#, Laura C. Cook#, Che-Chi Shu, Yuching Chen, Dawn Manias, Doraiswami Ramkrishna, Gary M. Dunny and Wei-Shou Hu* (2013), “Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer.” Proceedings of National Academy of Sciences, 110(17), 7086-7090.
- Anushree Chatterjee, Christopher M. Johnson, Che-Chi Shu, Yiannis N. Kaznessis, Doraiswami Ramkrishna, Gary M. Dunny and Wei-Shou Hu* (2011).“Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation.” Proceedings of the National Academy of Sciences, 108: 9721-9726.
- nushree Chatterjee, Laurie Drews, Sarika Mehra, Eriko Takano, Yiannis N. Kaznessis and Wei-Shou Hu* (2011),“Convergent transcription in the butyrolactone regulon in Streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis.” PLoS ONE, 6:e21974.
- Laetitia Canini, Jeremie Guedj, Anushree Chatterjee, Annabelle Lemenuel-Diot, Patrick F. Smith, and Alan S. Perelson* (2015), "Modeling the interaction between danoprevir and mericitabine in the treatment of chronic HCV infection." Antiviral Therapy (Advance article).
Che-Chi Shu, Anushree Chatterjee, Gary M. Dunny, Wei-Shou Hu and Doraiswami Ramkrishna* (2011), “Bistability versus Bimodal Distributions in Gene Regulatory Processes from Population Balance Model.” PLoS Computational Biology, 7(8):e1002140.
Laura CC Cook, Anushree Chatterjee, Aaron Barnes, Jeremy Yarwood, Wei-Shou Hu and GaryM. Dunny*(2011), “Biofilm growth alters regulation of conjugation by a bacterial pheromone.” Molecular Microbiology, 81(6), 1499-1510.
Che-Chi Shu,Anushree Chatterjee, Wei-Shou Hu and Doraiswami Ramkrishna* (2012), “Modeling of Gene regulatory processes by Population mediated Signaling. New applications of Population balances.” Chemical Engineering Science, 70: 188-199. (Link)

