Home
Welcome to the Spencer Lab!
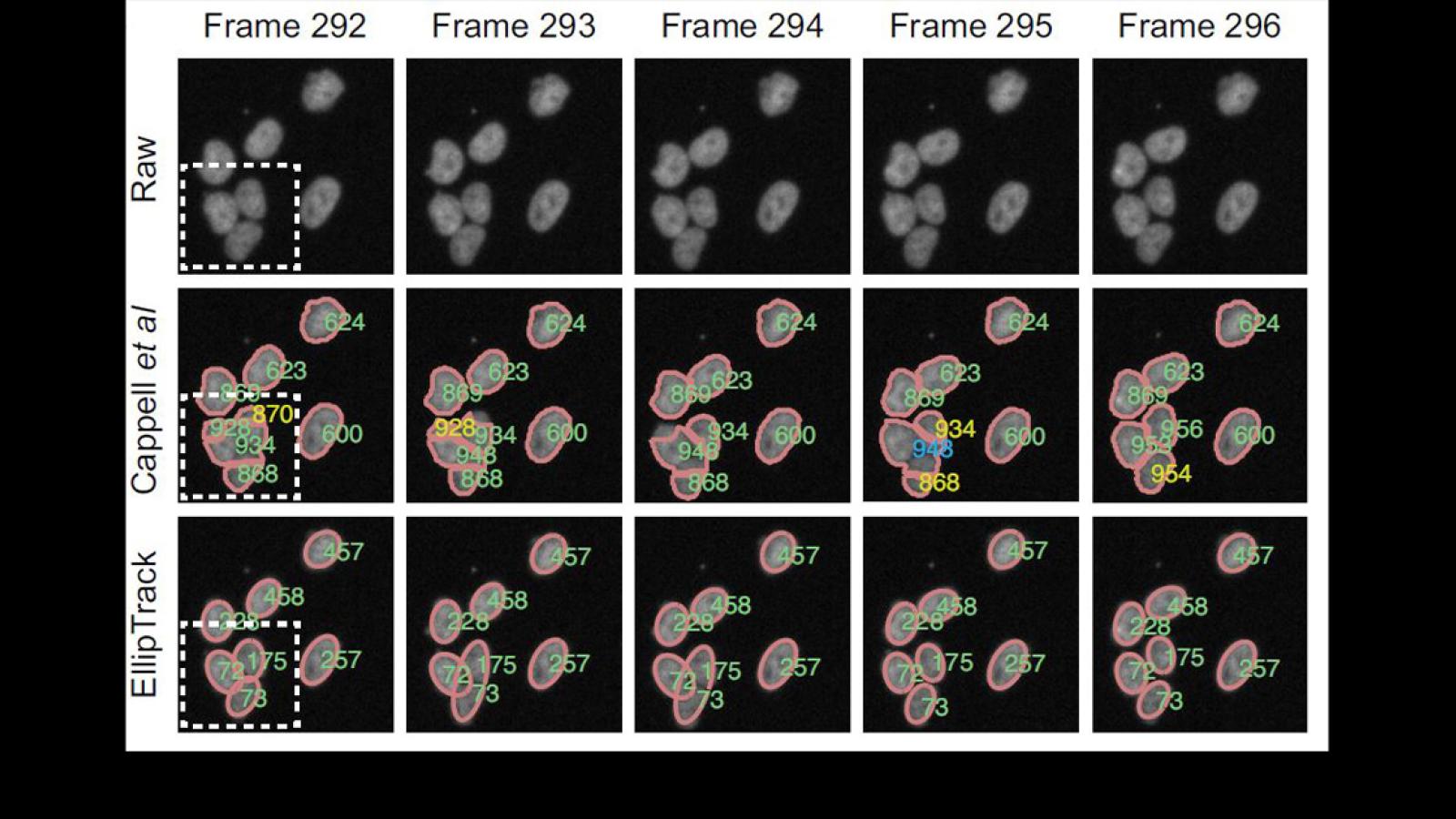
Research in the lab is focused on understanding how signaling events control cell fate. Studying these processes in single cells reveals remarkable cell-to-cell variability in response to stimuli, even among genetically identical cells in a uniform environment. We seek to understand the sources and consequences of this heterogeneity in the cellular response to stimuli. The stimuli we study include growth factors, cell stress, and targeted cancer therapeutics. To do this, we develop genetically encoded fluorescent sensors for signaling events of interest. We then use long-term live-cell microscopy and cell tracking to quantify the dynamics of upstream signals and link them to cell fate (proliferation, quiescence, apoptosis, differentiation). Our long-term goal is to understand the normal mechanistic functioning of signaling pathways that control proliferation, to understand how these signals go awry in cancer, and eventually to alter the fate of individual cells.