Research
Single-Molecule Imaging
Total internal reflection fluorescence microscopy (TIRFM)
Molecular Transport at Interfaces
The dynamic behavior of molecules and nanoparticles at
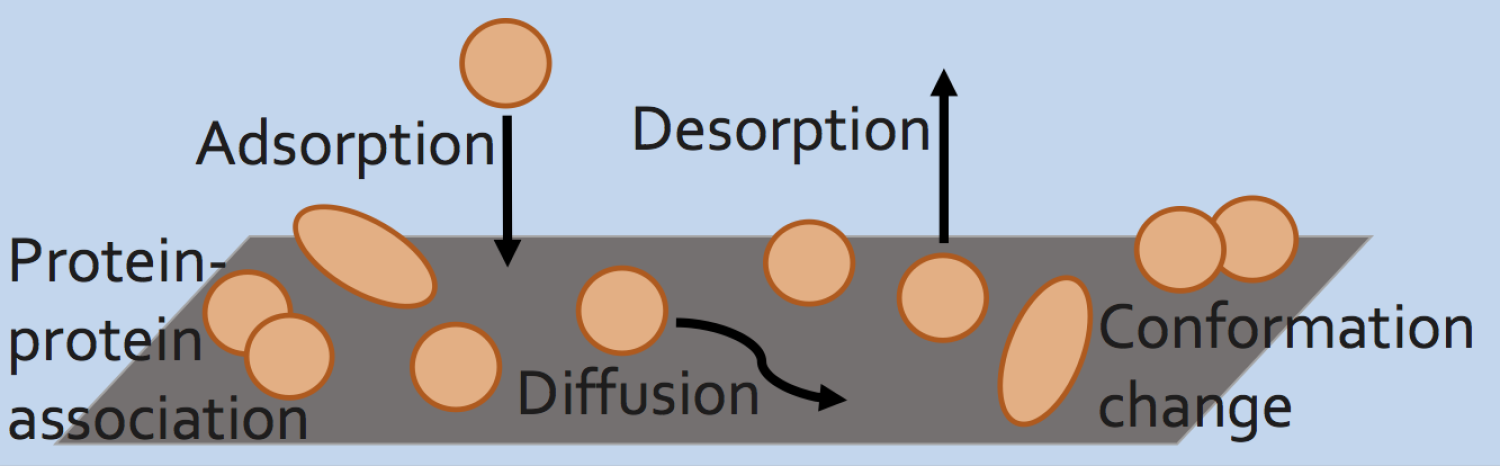
Mapping Using Accumulated Probe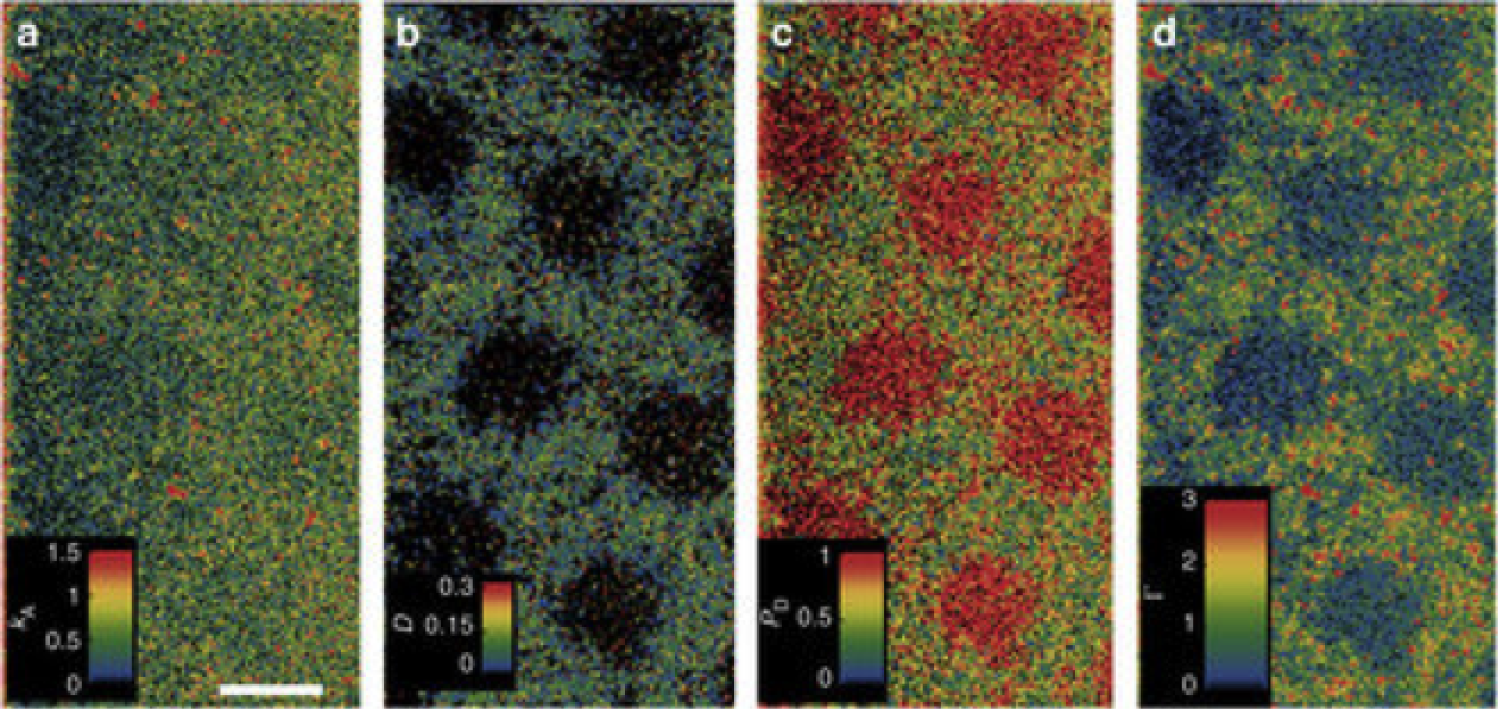 Trajectories (MAPT)
Trajectories (MAPT)

In MAPT, a time series of images is obtained using single-molecule TIFRM, and imaging processing is used to determine the locations of all the individual molecules. By tracking these locations through a time series, physical properties of probe/surface interactions (such as adsorption rates, local diffusion, desorption probability and surface coverage) are determined. By distributing these events to appropriate locations on the surface, one can determine characteristic values as a function of position. The use of single-molecule localization makes the MAPT analysis intrinsically super-resolution and we have demonstrated resolution of ~100 nm in our un-optimized initial results. In contrast with all previous super-resolution methods, MAPT can be used on any surface and provides absolute quantitative values of physical interaction properties. Homogenous surfaces can then be characterized and used as “calibration” points to connect small scale heterogeneities to specific surface chemistries. Using such maps, we found the first experimental evidence for desorption-mediated diffusion at the solid-liquid interface.
MAPT images of a photopatterned surface using multiple modes of contrast. (a) adsorption rate (1013 µm-2s-1M-1) (b) average diffusion coefficient (µm2/s) (c) desorption probability per 400 ms and (d) surface coverage/occupancy (1013 µm-2s-1M-1).
Amyloidogenic Protein Refolding in 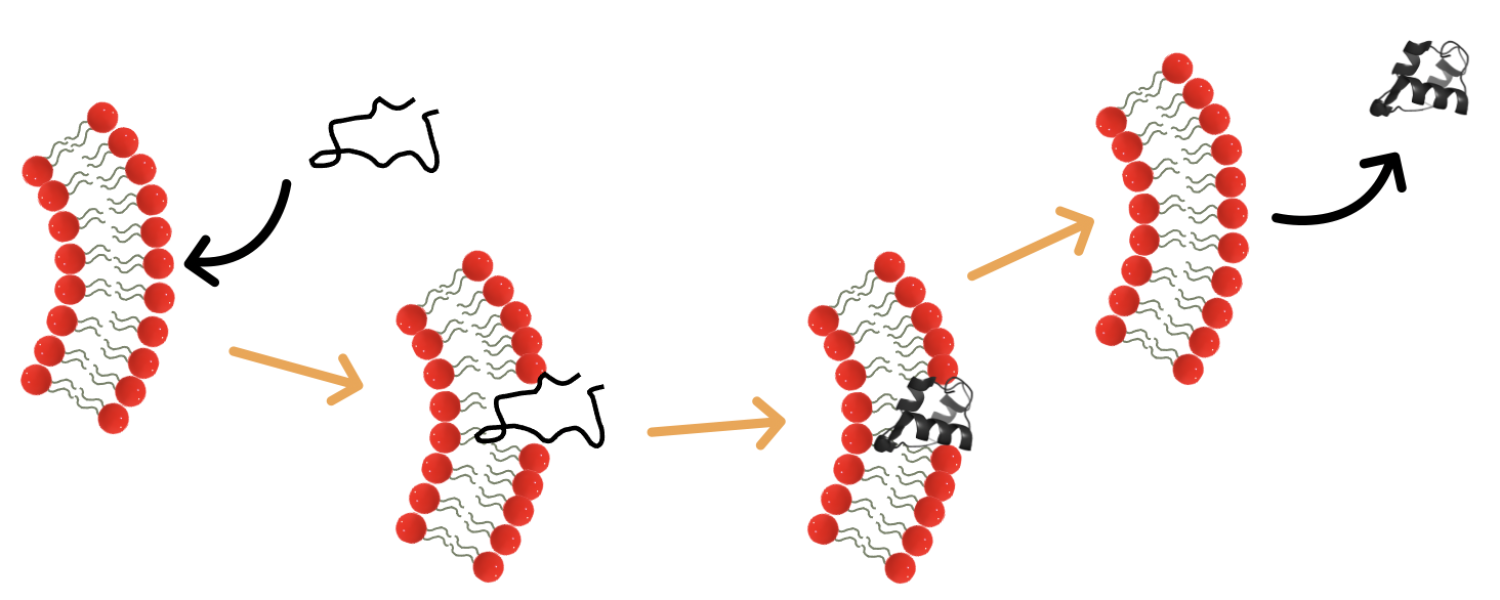 Lipidic and Polymeric Environments
Lipidic and Polymeric Environments

Amyloid fibrils are highly stable protein species that are correlated to numerous diseases, including Alzheimer's and Parkinson's. In the Schwartz Lab, we utilize functional biomaterials to stabilize amyloidogenic proteins, inhibit their fibrillization, and in some cases, even completely reverse that fibrillization. These functional materials, namely lipid vesicles and polymer brush functionalized nanoparticles, can be tuned to be optimally stabilizing for specific proteins. Numerous techniques including fluorescence spectroscopy, circular dichroism spectroscopy, and single molecule methods give insight into how these materials work.
Dynamics in Confined Environments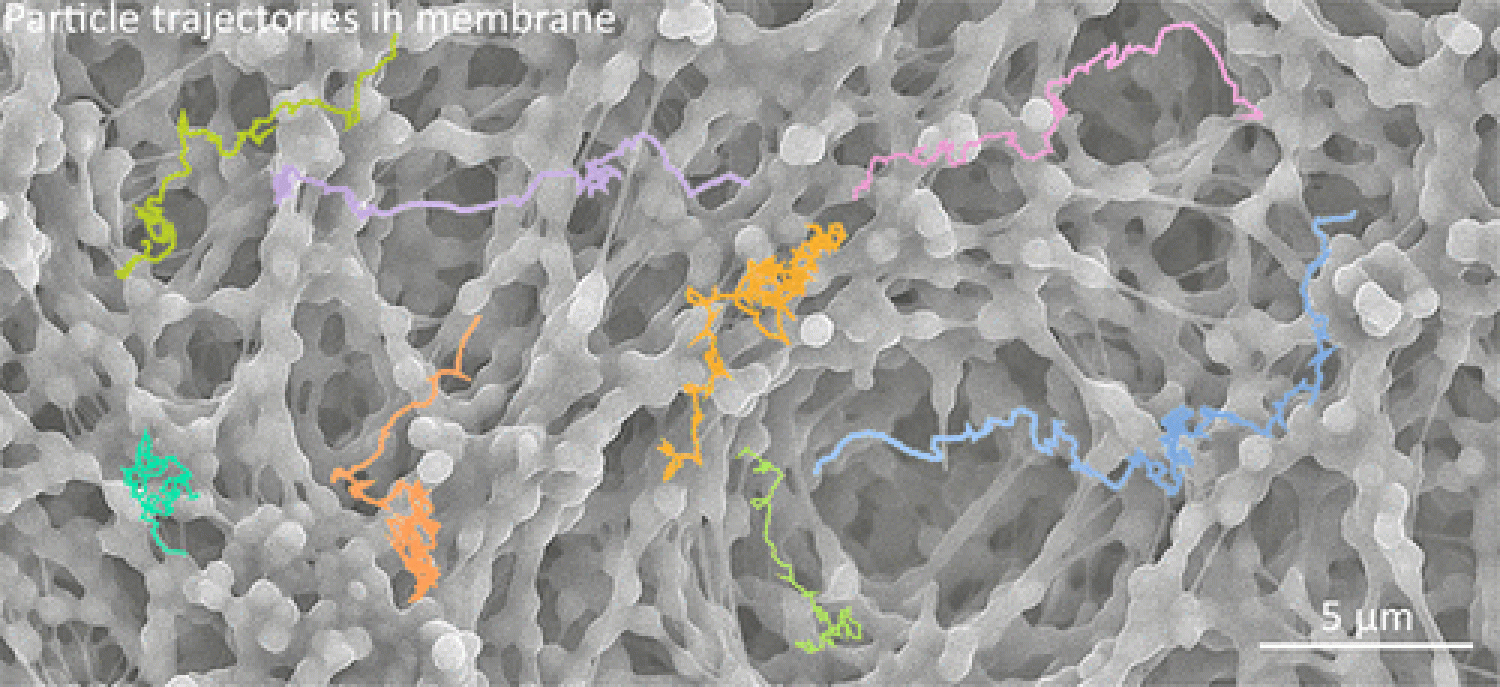

Understanding molecular behavior under confinement is critical to various applications including biosensing, material synthesis, intracellular and extracellular transport, and a number of industrial separation processes. In order to understand these environments, we have investigated systems ranging from surface crowding at the solid-liquid interface to structurally-induced confinement within synthesized materials. Based on single molecule trajectories, we have characterized dynamics using a variety of empirical and model dependent analyses. For example, it was discovered that poly(ethylene glycol) chains exhibited behavior consistent with the generalized continuous-time random walk model over a large range of surface coverage, from the dilute to well above the surface overlap concentration. Additionally, the polymer molecules had three distinct regimes of dynamics as a function of surface crowding, as shown by measurements of short-time diffusion coefficients, mean squared displacement, and surface waiting times. In other experiments, nanoparticle trajectories in porous polymer membranes were analyzed in order to measure the tortuosity of the membrane, which was found to be a function of the operating conditions and not merely a constant value. Moreover, distributions of the tortuosity indicated a large degree of spatiotemporal heterogeneity within the membranes, where anomalous, highly confined regions appeared to exist. Further experiments investigating dynamics in inverse opal structures are currently underway, where individual particle trajectories are being investigated in the context of the narrow escape problem in order to provide a better understanding of confined diffusion in periodic nanostructures.
Molecular Modification of Heterogeneous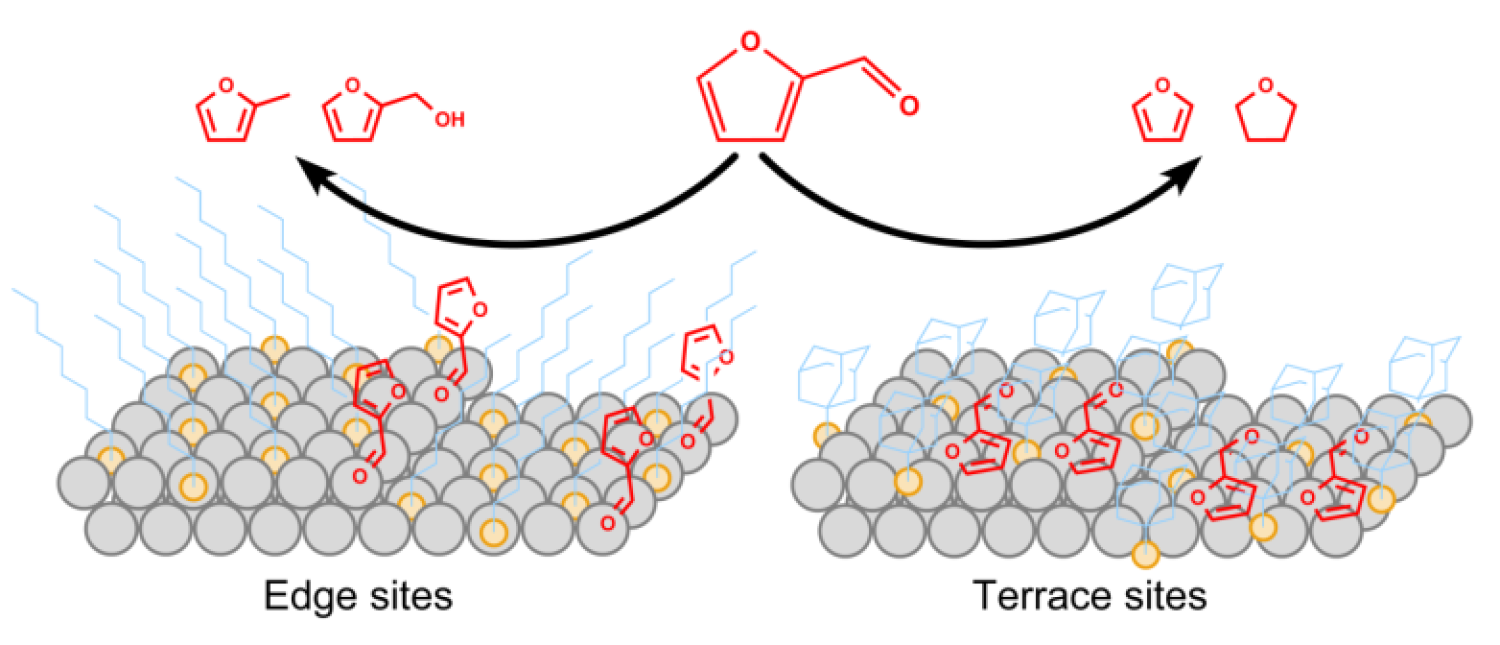 Catalysts
Catalysts

Performing selective conversions of reagents with multiple functional groups is a challenging objective since each group can potentially adsorb and react on a catalytic surface. One approach involves the modification of supported metal catalysts with organic ligands such as alkanethiols. Alkanethiols can be deposited on metal surfaces to form organized self-assembled monolayers (SAMs) that can greatly alter surface chemistry and potentially be used to significantly enhance selectivity of traditional catalytic systems. We have recently shown that such a strategy can be applied to technical supported catalysts such as Pd/Al2O3. This attachment strategy was found to dramatically enhance the selectivity to the desired products during furfural hydrogenation from < 5% on uncoated catalysts to > 90% on SAM coated catalysts by selectively blocking sites associated with the undesirable reaction pathway. By understanding how these ligands interact both with the surface as well as the reactants, we aim to tune their molecular structure to promote specific interactions for enhancing product selectivity.
Instruments and Methods
- Single-molecule imaging
- Total Internal Reflection Fluorescence Microscopy (TIRFM)
- Förster Resonance Energy Transfer (FRET)
- Double-helix point spread function
- Dual excitation
- Polarized light microscopy
- Surface modification (thin films and self-assembled monolayers)
- Surface characterization (contact angle goniometry, FTIR, ellipsometry)
- Atomic force microscopy
- Spectroscopy (circular dichroism, UV-vis, fluorescence)
- Numerical simulations and machine-learning
- Langmuir trough
- Brewster angle microscopy

