Comparison of explicit and mean-field models of cytoskeletal filaments with crosslinking motors
Adam R. Lamson, Jeffrey M. Moore, Fang Fang, Matthew A. Glaser, Michael J Shelley, and Meredith D. Betterton (2021). Eur. Phys. J. E. 44, 45. arXiv DOI: 2011.08156. DOI: 10.1140/epje/s10189-021-00042-9. Download.
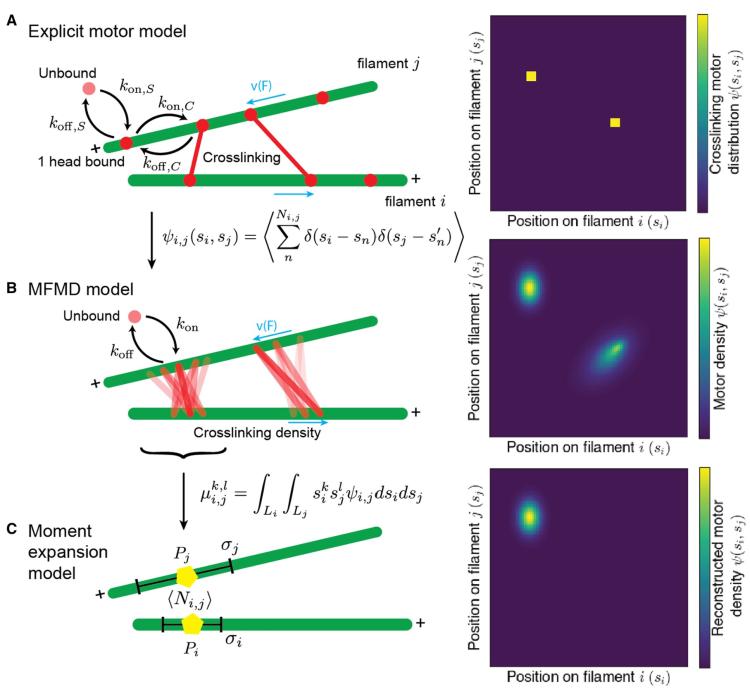
In cells, cytoskeletal filament networks are responsible for cell movement, growth, and division. Filaments in the cytoskeleton are driven and organized by crosslinking molecular motors. In reconstituted cytoskeletal systems, motor activity is responsible for far-from-equilibrium phenomena such as active stress, self-organized flow, and spontaneous nematic defect generation. How microscopic interactions between motors and filaments lead to larger-scale dynamics remains incompletely understood. To build from motor–filament interactions to predict bulk behavior of cytoskeletal systems, more computationally efficient techniques for modeling motor–filament interactions are needed. Here, we derive a coarse-graining hierarchy of explicit and continuum models for crosslinking motors that bind to and walk on filament pairs. We compare the steady-state motor distribution and motor-induced filament motion for the different models and analyze their computational cost. All three models agree well in the limit of fast motor binding kinetics. Evolving a truncated moment expansion of motor density speeds the computation by 103-106 compared to the explicit or continuous-density simulations, suggesting an approach for more efficient simulation of large networks. These tools facilitate further study of motor–filament networks on micrometer to millimeter length scales.

