Mechanisms of chromosome biorientation and bipolar spindle assembly analyzed by computational modeling
Christopher J. Edelmaier, Adam R. Lamson, Zachary R. Gergely, Saad Ansari, Robert Blackwell, J. Richard McIntosh, Matthew A. Glaser, and Meredith D. Betterton (2020). eLife9, e48787. Biorxiv DOI: 10.1101/649913. DOI: 10.7554/eLife.48787. Download.
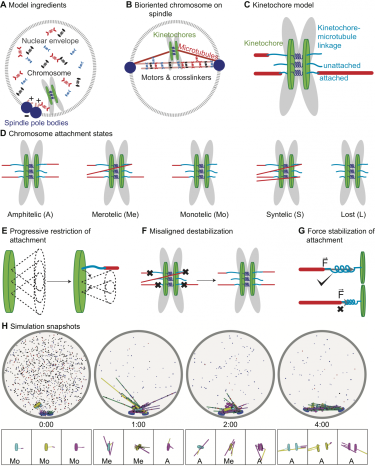
The essential functions required for mitotic spindle assembly and chromosome biorientation and segregation are not fully understood, despite extensive study. To illuminate the combinations of ingredients most important to align and segregate chromosomes and simultaneously assemble a bipolar spindle, we developed a computational model of fission-yeast mitosis. Robust chromosome biorientation requires progressive restriction of attachment geometry, destabilization of misaligned attachments, and attachment force dependence. Large spindle length fluctuations can occur when the kinetochore-microtubule attachment lifetime is long. The primary spindle force generators are kinesin-5 motors and crosslinkers in early mitosis, while interkinetochore stretch becomes important after biorientation. The same mechanisms that contribute to persistent biorientation lead to segregation of chromosomes to the poles after anaphase onset. This model therefore provides a framework to interrogate key requirements for robust chromosome biorientation, spindle length regulation, and force generation in the spindle.

