Using single molecule approaches to investigate mechanisms of transcription and formation of nucleoprotein complexes
Breadcrumb
The goal of this line of research is to investigate the assembly mechanism, dynamics, and heterogeneity of human transcription factor/DNA complexes, and how these parameters contribute to transcriptional control. To do so, we are leveraging the unique abilities of single molecule fluorescence microscopy, which provide unprecedented insight into how nucleoprotein complexes function. Single molecule experiments complement the knowledge gained from ensemble biochemical experiments by allowing the observation of sub-populations of molecules that exist in distinct states and also the measurement of dynamic behavior in individual molecules, which are obscured by averaging across all the molecules present in an ensemble.
We are interested in understanding the mechanisms governing interactions between proteins that are important for regulating transcription and the sites to which they bind in DNA and chromatin. Our studies primarily focus on transcriptional activator proteins and architectural proteins. For example, we have established systems to visualize single molecules of the transcriptional activator p53 and the architectural protein HMGB1 rapidly binding to nucleosomes, as well as scanning across a dense surface of nucleosomes. Our goal is to unravel how these binding modes occur and control the ability of these proteins to interact with chromatin.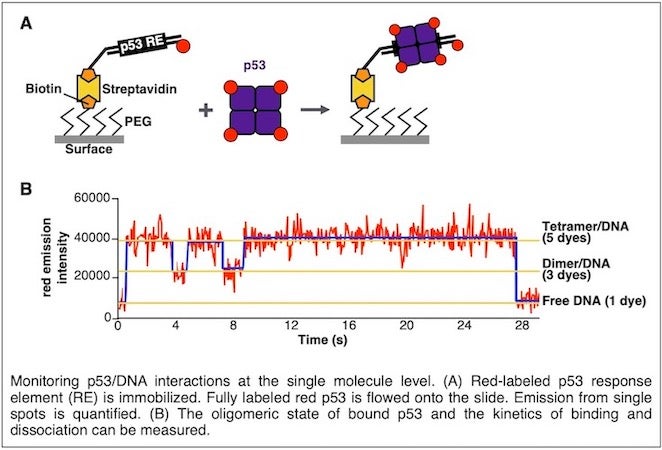
We are also investigating mechanisms of the transcription reaction itself. The first step in RNA polymerase II (Pol II) transcription is the formation of a preinitiation complex (PIC), which entails the general transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) assembling with Pol II on core promoter DNA. Real-time single molecule fluorescence imaging, both in cells and in vitro, has begun to illuminate dynamic mechanisms of PIC formation and Pol II transcription. We have developed an in vitro single molecule fluorescence transcription system that we are using to define dynamic transformations within Pol II complexes that involve the general transcription factors and control the partitioning between active and inactive transcription complexes.
transcription is the formation of a preinitiation complex (PIC), which entails the general transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) assembling with Pol II on core promoter DNA. Real-time single molecule fluorescence imaging, both in cells and in vitro, has begun to illuminate dynamic mechanisms of PIC formation and Pol II transcription. We have developed an in vitro single molecule fluorescence transcription system that we are using to define dynamic transformations within Pol II complexes that involve the general transcription factors and control the partitioning between active and inactive transcription complexes.
