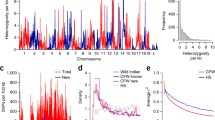
Abstract
We conducted whole-genome sequencing of four inbred mouse strains initially selected for high (H1, H2) or low (L1, L2) open-field activity (OFA), and then examined strain distribution patterns for all DNA variants that differed between their BALB/cJ and C57BL/6J parental strains. Next, we assessed genome-wide sharing (3,678,826 variants) both between and within the High and Low Activity strains. Results suggested that about 10% of these DNA variants may be associated with OFA, and clearly demonstrated its polygenic nature. Finally, we conducted bioinformatic analyses of functional genomics data from mouse, rat, and human to refine previously identified quantitative trait loci (QTL) for anxiety-related measures. This combination of sequence analysis and genomic-data integration facilitated refinement of previously intractable QTL findings, and identified possible genes for functional follow-up studies.




Similar content being viewed by others
Data availability
Raw reads from the 8 individuals sequenced have been deposited in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) with accession PRJNA605645.
References
Antoine MW, Vijayakumar S, McKeehan N, Jones SM, Hébert JM (2017) The severity of vestibular dysfunction in deafness as a determinant of comorbid hyperactivity or anxiety. J Neurosci Off J Soc Neurosci 37(20):5144–5154. https://doi.org/10.1523/JNEUROSCI.3545-16.2017
Anxiety and Depression Association of America, ADAA. Available at https://adaa.org/about-adaa/press-room/facts-statistics. Accessed 25 Sept 2019
Ashbrook DG, Williams RW, Lu L, Hager R (2015) A cross-species genetic analysis identifies candidate genes for mouse anxiety and human bipolar disorder. Front Behav Neurosci 9:171. https://doi.org/10.3389/fnbeh.2015.00171
Baker EJ, Jay JJ, Bubier JA, Langston MA, Chesler EJ (2011) GeneWeaver: a web-based system for integrative functional genomics. Nucleic Acids Res 40(D1):D1067–D1076. https://doi.org/10.1093/nar/gkr968
Bandelow B, Michaelis S, Wedekind D (2017) Treatment of anxiety disorders. Dialogues Clin Neurosci 19(2):93–107
Bloch-Zupan A, Sedano H, Scully C (2012) Dento/oro/craniofacial anomalies and genetics. Elsevier, Amsterdam. https://doi.org/10.1016/C2011-0-06093-3
Bockenhauer D, Feather S, Stanescu HC, Bandulik S, Zdebik AA, Reichold M, Tobin J, Lieberer E, Sterner C, Landoure G, Arora R, Sirimanna T, Thompson D, Cross JH, Van’t Hoff W, Al Masri O, Tullus K, Yeung S, Anikster Y, Klootwijk E, Hubank M, Dillon MJ, Heitzmann D, Arcos-Burgos M, Knepper MA, Dobbie A, Gahl W, Warth R, Sheridan E, Kleta R (2009) Epilepsy, ataxia, sensorineural deafness, tubulopathy, and KCNJ10 mutations. New Engl J Med 360(19):1960–1970. https://doi.org/10.1056/NEJMoa0810276
Bolivar V, Flaherty L (2003) A region on chromosome 15 controls intersession habituation in mice. J Neurosci 23(28):9435–9438. https://doi.org/10.1523/JNEUROSCI.23-28-09435.2003
Bubier JA, Wilcox TD, Jay JJ, Langston MA, Baker EJ, Chesler EJ (2016) Cross-species integrative functional genomics in GeneWeaver reveals a role for Pafah1b1 in altered response to alcohol. Front Behav Neurosci 10:1. https://doi.org/10.3389/fnbeh.2016.00001
Chapman EJ, Knowles MA (2009) Necdin: A multi functional protein with potential tumor suppressor role? Working Hypothesis 48(11). https://doi.org/10.1002/mc.20567
Chen Y, Zhou H, An Y, Jia X, Sun T, Guan X, Zhang J (2019) Combined effects of olfactory dysfunction and chronic stress on anxiety- and depressive- like behaviors in mice. Neurosci Lett 692:143–149. https://doi.org/10.1016/j.neulet.2018.11.010
Danecek P, Schiffels S, Durbin R (2014) Multiallelic calling model in bcftools (-m). 2
DeFries JC, Gervais MC, Thomas EA (1978) Response to 30 generations of selection for open-field activity in laboratory mice. Behav Genet 8(1):3–13. https://doi.org/10.1007/BF01067700
Devane CL, Chiao E, Franklin M, Kruep EJ (2005) Anxiety disorders in the 21st century: status, challenges, opportunities, and comorbidity with depression. Am J Manag Care 11(12 Suppl):S344–353
Duval ER, Javanbakht A, Liberzon I (2015) Neural circuits in anxiety and stress disorders: a focused review. Ther Clin Risk Manag 11:115–126. https://doi.org/10.2147/TCRM.S48528
Eser D, Leicht G, Lutz J, Wenninger S, Kirsch V, Schüle C, Karch S, Baghai T, Pogarell O, Born C, Rupprecht R, Mulert C (2009) Functional neuroanatomy of CCK-4-induced panic attacks in healthy volunteers. Hum Brain Mapp 30(2):511–522. https://doi.org/10.1002/hbm.20522
Fan K, Wu X, Fan B, Li N, Lin Y, Yao Y, Ma J (2012) Up-regulation of microglial cathepsin C expression and activity in lipopolysaccharide-induced neuroinflammation. J Neuroinflamm 9:96. https://doi.org/10.1186/1742-2094-9-96
Flint J, Corley R, DeFries JC, Fulker DW, Gray JA, Miller S, Collins AC (1995) A simple genetic basis for a complex psychological trait in laboratory mice. Science (New York, N.Y.) 269(5229):1432–1435. https://doi.org/10.1126/science.7660127
Gonzales NM, Seo J, Hernandez Cordero AI, St Pierre CL, Gregory JS, Distler MG, Abney M, Canzar S, Lionikas A, Palmer AA (2018) Genome wide association analysis in a mouse advanced intercross line. Nat Commun 9(1):5162. https://doi.org/10.1038/s41467-018-07642-8
Gottschalk MG, Domschke K (2017) Genetics of generalized anxiety disorder and related traits. Dialogues Clin Neurosci 19(2):159–168
Hashioka S, Inoue K, Miyaoka T, Hayashida M, Wake R, Oh-Nishi A, Inagaki M (2019) The possible causal link of periodontitis to neuropsychiatric disorders: more than psychosocial mechanisms. Int J Mol Sci 20(15):3723. https://doi.org/10.3390/ijms20153723
Henderson ND, Turri MG, DeFries JC, Flint J (2004) QTL analysis of multiple behavioral measures of anxiety in mice. Behav Genet 34(3):267–293. https://doi.org/10.1023/B:BEGE.0000017872.25069.44
Jiao H, Hong W, Nevo E, Li K, Zhao H (2019) Convergent reduction of V1R genes in subterranean rodents. BMC Evol Biol 19(1):176. https://doi.org/10.1186/s12862-019-1502-4
Le-Niculescu H, Balaraman Y, Patel SD, Ayalew M, Gupta J, Kuczenski R, Shekhar A, Schork N, Geyer MA, Niculescu AB (2011) Convergent functional genomics of anxiety disorders: translational identification of genes, biomarkers, pathways and mechanisms. Transl Psychiatry 1(5):e9–e9. https://doi.org/10.1038/tp.2011.9
Levey DF, Gelernter J, Polimanti R, Zhou H, Cheng Z, Aslan M, Quaden R, Concato J, Radhakrishnan K, Bryois J, Sullivan PF, Stein MB (2020) Reproducible genetic risk loci for anxiety: results from ∼200,000 participants in the Million Veteran Program. Am J Psychiatry. https://doi.org/10.1176/appi.ajp.2019.19030256
Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics (Oxford, England) 27(21):2987–2993. https://doi.org/10.1093/bioinformatics/btr509
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv:1303.3997 [q-Bio]. https://arxiv.org/abs/1303.3997
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 25(14):1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics (Oxford, England) 25(16):2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Lo C-L, Lossie AC, Liang T, Liu Y, Xuei X, Lumeng L, Zhou FC, Muir WM (2016) High resolution genomic scans reveal genetic architecture controlling alcohol preference in bidirectionally selected rat model. PLoS Genet 12(8):e1006178. https://doi.org/10.1371/journal.pgen.1006178
Meier SM, Deckert J (2019) Genetics of anxiety disorders. Curr Psychiatry Rep 21(3):16. https://doi.org/10.1007/s11920-019-1002-7
Meier SM, Trontti K, Purves KL, Als TD, Grove J, Laine M, Pedersen MG, Bybjerg-Grauholm J, Bækved-Hansen M, Sokolowska E, Mortensen PB, Hougaard DM, Werge T, Nordentoft M, Breen G, Børglum AD, Eley TC, Hovatta I, Mattheisen M, Mors O (2019) Genetic variants associated with anxiety and stress-related disorders: a Genome-Wide Association Study and Mouse-Model Study. JAMA Psychiatry. https://doi.org/10.1001/jamapsychiatry.2019.1119
Mouse Map Converter. (n.d.). Available at https://churchill-lab.jax.org/mousemapconverter/. Accessed 9 Oct 2019
Mouse Genomes Project. Available at https://www.sanger.ac.uk/science/data/mouse-genomes-project. Accessed 1 Mar 2020
Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD (2009) PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res 38:D204–D210. https://doi.org/10.1093/nar/gkp1019
Miller AH, Raison CL (2016) The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nat Rev Immunol 16(1):22–34. https://doi.org/10.1038/nri.2015.5
Mozhui K, Ciobanu DC, Schikorski T, Wang X, Lu L, Williams RW (2008) Dissection of a QTL hotspot on mouse distal chromosome 1 that modulates neurobehavioral phenotypes and gene expression. PLoS Genet 4(11):e1000260. https://doi.org/10.1371/journal.pgen.1000260
NIMH » Any Anxiety Disorder. (n.d.). Available at https://www.nimh.nih.gov/health/statistics/any-anxiety-disorder.shtml. Accessed 25 Sept 2019
Parker CC, Carbonetto P, Sokoloff G, Park YJ, Abney M, Palmer AA (2014) High-resolution genetic mapping of complex traits from a combined analysis of F2 and advanced intercross mice. Genetics 198(1):103–116. https://doi.org/10.1534/genetics.114.167056
Peterlik D, Flor PJ, Uschold-Schmidt N (2016) The emerging role of metabotropic glutamate receptors in the pathophysiology of chronic stress-related disorders. Curr Neuropharmacol 14(5):514–539. https://doi.org/10.2174/1570159X13666150515234920
Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, Hamre KM, Lariviere WR, Matthews DB, Mittleman G, Goldowitz D, Chesler EJ (2010) High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes, Brain, Behav 9(2):129–159. https://doi.org/10.1111/j.1601-183X.2009.00540.x
Purves KL, Coleman JRI, Meier SM, Rayner C, Davis KAS, Cheesman R, Bækvad-Hansen M, Børglum AD, Wan Cho S, Jürgen Deckert J, Gaspar HA, Bybjerg-Grauholm J, Hettema JM, Hotopf M, Hougaard D, Hübel C, Kan C, McIntosh AM, Mors O et al (2019) A major role for common genetic variation in anxiety disorders. Mol Psychiatry. https://doi.org/10.1038/s41380-019-0559-1
Reference GH (n.d.). CTSC gene. Genetics Home Reference. Available at https://ghr.nlm.nih.gov/gene/CTSC. Accessed 5 Feb 2020
Reich D, Thangaraj K, Patterson N, Price AL, Singh L (2009) Reconstructing Indian population history. Nature 461(7263):489–494. https://doi.org/10.1038/nature08365
Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C (2003) Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301(5637):1211–1216. https://doi.org/10.1126/science.1086949
Seibenhener ML, Wooten MC (2015) Use of the open field maze to measure locomotor and anxiety-like behavior in mice. J Vis Exp JoVE. https://doi.org/10.3791/52434
Shimada-Sugimoto M, Otowa T, Hettema JM (2015) Genetics of anxiety disorders: genetic epidemiological and molecular studies in humans. Psychiatry Clin Neurosci 69(7):388–401. https://doi.org/10.1111/pcn.12291
Sokal RR, Rohlf FJ (1995) Biometry: The Principles and Practice of Statistics in Biological Research, 3rd edn. W.H. Freeman and Co., New York
Turri MG, Datta SR, DeFries J, Henderson ND, Flint J (2001a) QTL analysis identifies multiple behavioral dimensions in ethological tests of anxiety in laboratory mice. Curr Biol 11(10):725–734. https://doi.org/10.1016/s0960-9822(01)00206-8
Turri MG, Henderson ND, DeFries JC, Flint J (2001b) Quantitative trait locus mapping in laboratory mice derived from a replicated selection experiment for open-field activity. Genetics 158(3):1217–1226
Wadhawan A, Reynolds MA, Makkar H, Scott AJ, Potocki E, Hoisington AJ, Brenner LA, Dagdag A, Lowry CA, Dwivedi Y, Postolache TT (2020) Periodontal pathogens and neuropsychiatric health. Curr Top Med Chem. https://doi.org/10.2174/1568026620666200110161105
Ward J, Tunbridge EM, Sandor C, Lyall LM, Ferguson A, Strawbridge RJ, Lyall DM, Cullen B, Graham N, Johnston KJA, Webber C, Escott-Price V, O’Donovan M, Pell JP, Bailey MES, Harrison PJ, Smith DJ (2019) The genomic basis of mood instability: identification of 46 loci in 363,705 UK Biobank participants, genetic correlation with psychiatric disorders, and association with gene expression and function. Mol Psychiatry. https://doi.org/10.1038/s41380-019-0439-8
Waters RP, Pringle RB, Forster GL, Renner KJ, Malisch JL, Garland T Jr, Swallow JG (2013) Selection for increased voluntary wheel-running affects behavior and brain monoamines in mice. Brain Res 1508:9–22. https://doi.org/10.1016/j.brainres.2013.01.033
Zhou Z, Blandino P, Yuan Q, Shen P-H, Hodgkinson CA, Virkkunen M, Watson SJ, Akil H, Goldman D (2019) Exploratory locomotion, a predictor of addiction vulnerability, is oligogenic in rats selected for this phenotype. Proc Natl Acad Sci 116(26):13107–13115. https://doi.org/10.1073/pnas.1820410116
Acknowledgements
We would like to thank Dr. Erich J Baker for his assistance with GeneWeaver updates and functions.
Funding
This study was funded by NIDA and NIAAA (Grant No. R01 AA019776) and NIMH (Grant No. R01 MH100141-06). R01 AA018776 (NIH NIDA and NIAAA) to EJC. R01 MH100141-06 (NIMH to Matthew C. Keller) supports LME. R21 MH116263A (NIMH) supports CAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Aimee L. Thomas, Luke M. Evans, Michaela D. Nelsen, Elissa J. Chesler, Matthew S. Powers, Winona C. Booher, Christopher A. Lowry, John C. DeFries, Marissa A. Ehringer have no conflict of interest to declare.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Human and animal rights and informed consent
The local Institutional Animal Care and Use Committee (IACUC) approved all protocols using the guidelines set forth by the Office of Laboratory Animal Welfare. All animal protocols were approved by the IACUC and follow all international regulations for human treatment. No human subjects were involved in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thomas, A.L., Evans, L.M., Nelsen, M.D. et al. Whole-Genome Sequencing of Inbred Mouse Strains Selected for High and Low Open-Field Activity. Behav Genet 51, 68–81 (2021). https://doi.org/10.1007/s10519-020-10014-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10519-020-10014-y